Working with PDB Structures in Pandas
Let’s start by loading up a PDB structure from my favourite OPIG tool, the STCRDab! Here we will look at PDB ID 6eqb. It is a crystal structure of a T cell receptor interacting with a peptide presented by an MHC class I molecule.
[1]:
import nglview
[2]:
view = nglview.show_file('data/6eqb.pdb')
view
Now let’s load this molecules into a pandas dataframe and do some analysis. The PDB data is loaded into columns in a similar way that PDB files are formatted as columns.
[3]:
from python_pdb.parsers import parse_pdb_to_pandas
[4]:
with open('data/6eqb.pdb', 'r') as fh:
df = parse_pdb_to_pandas(fh.read())
df
[4]:
| record_type | atom_number | atom_name | alt_loc | residue_name | chain_id | residue_seq_id | residue_insert_code | pos_x | pos_y | pos_z | occupancy | b_factor | element | charge | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ATOM | 1 | N | A | ALA | C | 2 | None | 48.681 | -11.013 | 29.600 | 0.5 | 30.86 | N | None |
| 1 | ATOM | 2 | CA | A | ALA | C | 2 | None | 49.343 | -9.708 | 29.330 | 0.5 | 29.82 | C | None |
| 2 | ATOM | 3 | C | A | ALA | C | 2 | None | 49.310 | -8.792 | 30.562 | 0.5 | 29.16 | C | None |
| 3 | ATOM | 4 | O | A | ALA | C | 2 | None | 48.668 | -9.107 | 31.537 | 0.5 | 29.75 | O | None |
| 4 | ATOM | 5 | CB | A | ALA | C | 2 | None | 48.679 | -9.044 | 28.146 | 0.5 | 29.57 | C | None |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 6644 | HETATM | 6645 | O | None | HOH | B | 128 | None | 63.467 | -9.052 | 4.232 | 1.0 | 46.86 | O | None |
| 6645 | HETATM | 6646 | O | None | HOH | B | 129 | None | 64.998 | -3.887 | 10.735 | 1.0 | 48.05 | O | None |
| 6646 | HETATM | 6647 | O | None | HOH | B | 130 | None | 69.089 | -37.773 | 1.079 | 1.0 | 63.55 | O | None |
| 6647 | HETATM | 6648 | O | None | HOH | B | 131 | None | 70.137 | -40.643 | 1.067 | 1.0 | 59.75 | O | None |
| 6648 | HETATM | 6649 | O | None | HOH | B | 132 | None | 66.479 | 4.782 | 14.497 | 1.0 | 64.56 | O | None |
6649 rows × 15 columns
To start things off, let’s clean up this structure by highlighting one of the most powerful aspects of this approach: querying. As you can seen from the data frame above- there are water molecules in the structure that we might not care about. Let’s remove them…
[5]:
df_clean = df.query("record_type == 'ATOM'") # or "residue_name != 'HOH'" would have worked as well
df_clean.tail()
[5]:
| record_type | atom_number | atom_name | alt_loc | residue_name | chain_id | residue_seq_id | residue_insert_code | pos_x | pos_y | pos_z | occupancy | b_factor | element | charge | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6633 | ATOM | 6634 | CB | None | MET | B | 125 | None | 74.125 | -32.337 | 9.293 | 1.0 | 121.02 | C | None |
| 6634 | ATOM | 6635 | CG | None | MET | B | 125 | None | 73.143 | -31.632 | 10.263 | 1.0 | 116.33 | C | None |
| 6635 | ATOM | 6636 | SD | None | MET | B | 125 | None | 71.379 | -32.103 | 10.373 | 1.0 | 113.91 | S | None |
| 6636 | ATOM | 6637 | CE | None | MET | B | 125 | None | 70.626 | -31.028 | 9.134 | 1.0 | 109.06 | C | None |
| 6637 | ATOM | 6638 | OXT | None | MET | B | 125 | None | 74.839 | -34.197 | 6.246 | 1.0 | 130.70 | O | None |
By using pandas’ built in querying function- we can easily get rid of the HETATMs in the file that we might not care about. We can also use this querying to just select the TCR or pMHC to perform analysis on this molecule seperately. In this example, the TCR \(\alpha\)- and \(\beta\) chain is labelled as chains D and E repectively. The MHC molecule is chain A (B- representing the \(\beta_2\)m) and C the peptide.
[6]:
!head -n5 data/6eqb.pdb
REMARK 5 IMGT RENUMBERED STRUCTURE 6EQB GENERATED BY STCRDAB
REMARK 5 TCR CHAINS ARE RENUMBERED IN THE VARIABLE REGIONS ONLY
REMARK 5 MHC CHAINS ARE RENUMBERED IN THE G DOMAINS OR FOR B2M-GLOBULIN
REMARK 5 NON-TCR AND NON-MHC CHAINS ARE LEFT WITH RESIDUE IDS AS IN PDB
REMARK 5 PAIRED_ABTCR BCHAIN=E ACHAIN=D MHCCHAINS=AB AGCHAIN=C AGTYPE=PEPTIDE
[7]:
tcr_df = df_clean.query("chain_id == 'D' or chain_id == 'E'")
tcr_df
[7]:
| record_type | atom_number | atom_name | alt_loc | residue_name | chain_id | residue_seq_id | residue_insert_code | pos_x | pos_y | pos_z | occupancy | b_factor | element | charge | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 57 | ATOM | 58 | N | None | SER | E | 1 | None | 44.786 | 19.936 | 31.694 | 1.0 | 91.71 | N | None |
| 58 | ATOM | 59 | CA | None | SER | E | 1 | None | 43.328 | 20.085 | 31.879 | 1.0 | 88.14 | C | None |
| 59 | ATOM | 60 | C | None | SER | E | 1 | None | 42.582 | 19.234 | 30.860 | 1.0 | 80.31 | C | None |
| 60 | ATOM | 61 | O | None | SER | E | 1 | None | 41.944 | 19.762 | 29.940 | 1.0 | 81.48 | O | None |
| 61 | ATOM | 62 | CB | None | SER | E | 1 | None | 42.912 | 21.571 | 31.778 | 1.0 | 93.53 | C | None |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 3519 | ATOM | 3520 | CD1 | None | PHE | D | 215 | None | -5.784 | 33.229 | 67.928 | 1.0 | 154.28 | C | None |
| 3520 | ATOM | 3521 | CD2 | None | PHE | D | 215 | None | -6.202 | 32.551 | 65.649 | 1.0 | 145.03 | C | None |
| 3521 | ATOM | 3522 | CE1 | None | PHE | D | 215 | None | -6.684 | 34.292 | 67.786 | 1.0 | 160.08 | C | None |
| 3522 | ATOM | 3523 | CE2 | None | PHE | D | 215 | None | -7.092 | 33.613 | 65.506 | 1.0 | 151.52 | C | None |
| 3523 | ATOM | 3524 | CZ | None | PHE | D | 215 | None | -7.336 | 34.483 | 66.576 | 1.0 | 159.15 | C | None |
3461 rows × 15 columns
[8]:
mhc_df = df_clean.query("chain_id == 'A'")
mhc_df
[8]:
| record_type | atom_number | atom_name | alt_loc | residue_name | chain_id | residue_seq_id | residue_insert_code | pos_x | pos_y | pos_z | occupancy | b_factor | element | charge | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3541 | ATOM | 3542 | N | None | GLY | A | 1 | None | 63.937 | -26.599 | 37.997 | 1.0 | 88.67 | N | None |
| 3542 | ATOM | 3543 | CA | None | GLY | A | 1 | None | 64.891 | -25.601 | 38.583 | 1.0 | 88.18 | C | None |
| 3543 | ATOM | 3544 | C | None | GLY | A | 1 | None | 64.208 | -24.275 | 38.448 | 1.0 | 82.85 | C | None |
| 3544 | ATOM | 3545 | O | None | GLY | A | 1 | None | 63.079 | -24.143 | 38.876 | 1.0 | 81.66 | O | None |
| 3545 | ATOM | 3546 | N | None | SER | A | 2 | None | 64.856 | -23.315 | 37.808 | 1.0 | 80.55 | N | None |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 5790 | ATOM | 5791 | O | None | PRO | A | 1186 | None | 77.326 | -47.026 | 15.302 | 1.0 | 147.25 | O | None |
| 5791 | ATOM | 5792 | CB | None | PRO | A | 1186 | None | 79.797 | -49.588 | 14.420 | 1.0 | 159.44 | C | None |
| 5792 | ATOM | 5793 | CG | None | PRO | A | 1186 | None | 81.295 | -49.731 | 14.585 | 1.0 | 160.29 | C | None |
| 5793 | ATOM | 5794 | CD | None | PRO | A | 1186 | None | 81.803 | -48.494 | 15.300 | 1.0 | 158.37 | C | None |
| 5794 | ATOM | 5795 | OXT | None | PRO | A | 1186 | None | 77.441 | -48.931 | 16.082 | 1.0 | 151.45 | O | None |
2254 rows × 15 columns
[9]:
peptide_df = df_clean.query("chain_id == 'C'")
peptide_residues_df = peptide_df.groupby(['residue_seq_id', 'residue_insert_code'], dropna=False)
print(f'The peptide is a {len(peptide_residues_df)}-mer!')
The peptide is a 9-mer!
Another advantage of using pandas is we can add new columns to annotate properties in the structure that we care about. In this example, since the TCR is from STCRDab and has been renumbered using the IMGT numbering convention we can easily identify the complimentary determining regions based on their residue_seq_id property.
[10]:
IMGT_CDR1 = set(range(27, 38 + 1))
IMGT_CDR2 = set(range(56, 65 + 1))
IMGT_CDR3 = set(range(105, 117 + 1))
def assign_cdr_number(seq_id: int) -> int | None:
'''
Map imgt_id to CDR domains, return number associated with domain or return None if input is not in a CDR
domain.
'''
if seq_id in IMGT_CDR1:
return 1
if seq_id in IMGT_CDR2:
return 2
if seq_id in IMGT_CDR3:
return 3
return None
tcr_df = tcr_df.copy() # Doing this on a copy of the dataframe since it is originally a slice of df!
tcr_df['cdr'] = tcr_df['residue_seq_id'].map(assign_cdr_number)
We can also annotations for the \(\alpha\) and \(\beta\) chain since these are defined by the STCRDab header.
[11]:
tcr_df['chain_type'] = tcr_df['chain_id'].map(lambda chain_id: 'alpha' if chain_id == 'D' else 'beta')
Now we can easily get rich information about the TCR CDR loops with ease.
[12]:
tcr_cdrs_df = tcr_df.query('cdr.notnull()')
cdr_lengths = tcr_cdrs_df[
['chain_type', 'cdr', 'residue_seq_id', 'residue_insert_code']
].drop_duplicates().groupby(['chain_type', 'cdr'], dropna=False).size()
cdr_lengths
[12]:
chain_type cdr
alpha 1.0 6
2.0 6
3.0 9
beta 1.0 6
2.0 5
3.0 13
dtype: int64
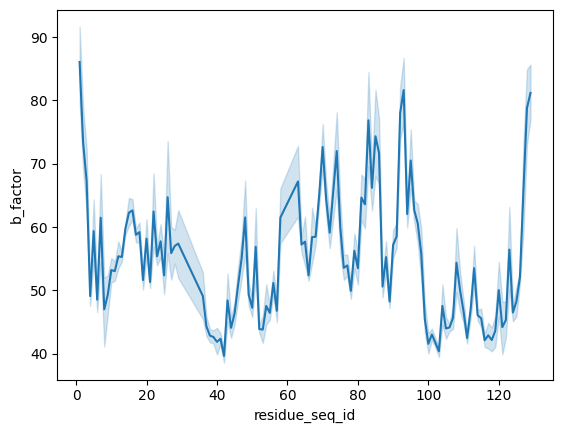
We can also easily properties such as b-factors in the TCR variable domain.
[13]:
import seaborn as sns
[14]:
tcr_variable_df = tcr_df.query('residue_seq_id <= 129')
sns.lineplot(x=tcr_variable_df['residue_seq_id'], y=tcr_variable_df['b_factor'])
[14]:
<Axes: xlabel='residue_seq_id', ylabel='b_factor'>
Computing things like contacting residues between the TCR CDR loops and the peptide is a breeze.
[15]:
import numpy as np
[16]:
CONTACT_DISTANCE = 5 # Angstroms (Å)
def euclidean_distance(x1, y1, z1, x2, y2, z2):
return np.sqrt((x2 - x1)**2 + (y2 - y1)**2 + (z2 - z1)**2)
interface = tcr_cdrs_df.merge(peptide_df, how='cross', suffixes=('_tcr', '_peptide'))
interface['atom_distances'] = euclidean_distance(interface['pos_x_tcr'], interface['pos_y_tcr'], interface['pos_z_tcr'],
interface['pos_x_peptide'], interface['pos_y_peptide'], interface['pos_z_peptide'])
contacting_atoms = interface[interface['atom_distances'] <= CONTACT_DISTANCE]
contacting_residues = contacting_atoms[['chain_id_tcr', 'residue_seq_id_tcr', 'residue_insert_code_tcr', 'cdr', 'chain_type',
'residue_seq_id_peptide', 'residue_insert_code_peptide']].drop_duplicates()
contacting_residues
[16]:
| chain_id_tcr | residue_seq_id_tcr | residue_insert_code_tcr | cdr | chain_type | residue_seq_id_peptide | residue_insert_code_peptide | |
|---|---|---|---|---|---|---|---|
| 6424 | E | 108 | None | 3.0 | beta | 8 | None |
| 6603 | E | 109 | None | 3.0 | beta | 9 | None |
| 6641 | E | 109 | None | 3.0 | beta | 7 | None |
| 6704 | E | 109 | None | 3.0 | beta | 8 | None |
| 6926 | E | 110 | None | 3.0 | beta | 7 | None |
| 7151 | E | 111 | None | 3.0 | beta | 7 | None |
| 7194 | E | 111 | None | 3.0 | beta | 4 | None |
| 7253 | E | 111 | None | 3.0 | beta | 5 | None |
| 7261 | E | 111 | None | 3.0 | beta | 6 | None |
| 7280 | E | 111 | None | 3.0 | beta | 8 | None |
| 7361 | E | 111 | None | 3.0 | beta | 3 | None |
| 7594 | E | 112 | None | 3.0 | beta | 4 | None |
| 7596 | E | 112 | None | 3.0 | beta | 5 | None |
| 7603 | E | 112 | None | 3.0 | beta | 6 | None |
| 7607 | E | 112 | None | 3.0 | beta | 7 | None |
| 7622 | E | 112 | None | 3.0 | beta | 8 | None |
| 8114 | E | 113 | None | 3.0 | beta | 5 | None |
| 11763 | D | 37 | None | 1.0 | alpha | 5 | None |
| 11923 | D | 37 | None | 1.0 | alpha | 4 | None |
| 12030 | D | 37 | None | 1.0 | alpha | 2 | None |
| 12033 | D | 37 | None | 1.0 | alpha | 3 | None |
| 12218 | D | 38 | None | 1.0 | alpha | 5 | None |
| 13131 | D | 57 | None | 2.0 | alpha | 5 | None |
Finally, if we want to save part of the data frame as a PDB file, we can convert it back to a Structure object and save it to a file.
[17]:
import warnings
from python_pdb.entities import Structure, StructureConstructionWarning
[18]:
# Suppressing warnings here because there are alternate locations specified in this PDB file
with warnings.catch_warnings():
warnings.filterwarnings('ignore', category=StructureConstructionWarning)
tcr_structure = Structure.from_pandas(tcr_df)
with open('tcr.pdb', 'w') as fh:
fh.write(str(tcr_structure))
[19]:
!ls | grep '.pdb'
tcr.pdb
[20]:
!head tcr.pdb
ATOM 58 N SER E 1 44.786 19.936 31.694 1.00 91.71 N
ATOM 59 CA SER E 1 43.328 20.085 31.879 1.00 88.14 C
ATOM 60 C SER E 1 42.582 19.234 30.860 1.00 80.31 C
ATOM 61 O SER E 1 41.944 19.762 29.940 1.00 81.48 O
ATOM 62 CB SER E 1 42.912 21.571 31.778 1.00 93.53 C
ATOM 63 OG SER E 1 41.483 21.690 31.841 1.00 93.62 O
ATOM 64 N GLN E 2 42.634 17.921 31.043 1.00 72.67 N
ATOM 65 CA GLN E 2 41.716 17.039 30.308 1.00 69.30 C
ATOM 66 C GLN E 2 40.246 17.307 30.674 1.00 66.83 C
ATOM 67 O GLN E 2 39.999 17.911 31.668 1.00 70.99 O