Centre of mass analysis
Introduction
In this notebook we aimed to identify the centre-of-mass (COM) of the TCR framework (Fw) regions and the anchors of the CDR loops. We then investigated the amount of movement these regions undergo between apo and holo states. We also used these COMs to create an axis of the TCR and measure the angle between both chains of the TCR before determining the angle changes between apo and holo states.
[1]:
import os
from collections import defaultdict
import numpy as np
import pandas as pd
import seaborn as sns
from python_pdb.parsers import parse_pdb_to_pandas
from tcr_pmhc_interface_analysis.measurements import get_distance
from tcr_pmhc_interface_analysis.processing import annotate_tcr_pmhc_df, find_anchors
from tcr_pmhc_interface_analysis.utils import get_coords
[2]:
DATA_DIR = '../data/processed/apo-holo-tcr-pmhc-class-I'
[3]:
apo_holo_summary_df = pd.read_csv(os.path.join(DATA_DIR, 'apo_holo_summary.csv'))
apo_holo_summary_df
[3]:
| file_name | pdb_id | structure_type | state | alpha_chain | beta_chain | antigen_chain | mhc_chain1 | mhc_chain2 | cdr_sequences_collated | peptide_sequence | mhc_slug | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1ao7_D-E-C-A-B_tcr_pmhc.pdb | 1ao7 | tcr_pmhc | holo | D | E | C | A | B | DRGSQS-IYSNGD-AVTTDSWGKLQ-MNHEY-SVGAGI-ASRPGLA... | LLFGYPVYV | hla_a_02_01 |
| 1 | 1b0g_C-A-B_pmhc.pdb | 1b0g | pmhc | apo | NaN | NaN | C | A | B | NaN | ALWGFFPVL | hla_a_02_01 |
| 2 | 1b0g_F-D-E_pmhc.pdb | 1b0g | pmhc | apo | NaN | NaN | F | D | E | NaN | ALWGFFPVL | hla_a_02_01 |
| 3 | 1bd2_D-E-C-A-B_tcr_pmhc.pdb | 1bd2 | tcr_pmhc | holo | D | E | C | A | B | NSMFDY-ISSIKDK-AAMEGAQKLV-MNHEY-SVGAGI-ASSYPGG... | LLFGYPVYV | hla_a_02_01 |
| 4 | 1bii_P-A-B_pmhc.pdb | 1bii | pmhc | apo | NaN | NaN | P | A | B | NaN | RGPGRAFVTI | h2_dd |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 386 | 7rtd_C-A-B_pmhc.pdb | 7rtd | pmhc | apo | NaN | NaN | C | A | B | NaN | YLQPRTFLL | hla_a_02_01 |
| 387 | 7rtr_D-E-C-A-B_tcr_pmhc.pdb | 7rtr | tcr_pmhc | holo | D | E | C | A | B | DRGSQS-IYSNGD-AVNRDDKII-SEHNR-FQNEAQ-ASSPDIEQY | YLQPRTFLL | hla_a_02_01 |
| 388 | 8gvb_A-B-P-H-L_tcr_pmhc.pdb | 8gvb | tcr_pmhc | holo | A | B | P | H | L | YGATPY-YFSGDTLV-AVGFTGGGNKLT-SEHNR-FQNEAQ-ASSD... | RYPLTFGW | hla_a_24_02 |
| 389 | 8gvg_A-B-P-H-L_tcr_pmhc.pdb | 8gvg | tcr_pmhc | holo | A | B | P | H | L | YGATPY-YFSGDTLV-AVGFTGGGNKLT-SEHNR-FQNEAQ-ASSD... | RFPLTFGW | hla_a_24_02 |
| 390 | 8gvi_A-B-P-H-L_tcr_pmhc.pdb | 8gvi | tcr_pmhc | holo | A | B | P | H | L | YGATPY-YFSGDTLV-AVVFTGGGNKLT-SEHNR-FQNEAQ-ASSL... | RYPLTFGW | hla_a_24_02 |
391 rows × 12 columns
[4]:
complexes = [complex_id for complex_id in os.listdir(DATA_DIR)
if os.path.isdir(os.path.join(DATA_DIR, complex_id))]
Computing Measurements
[5]:
def calculate_angle(vec1, vec2):
'''Caculate the angle between two vectors.'''
return np.arccos(np.dot(vec1, vec2) / (np.linalg.norm(vec1) * np.linalg.norm(vec2)))
[6]:
complex_ids = []
structure_x_paths = []
structure_y_paths = []
alpha_anchor_com_diffs = []
beta_anchor_com_diffs = []
alpha_fw_com_diffs = []
beta_fw_com_diffs = []
chain_angle_diffs = []
for complex_id in complexes:
complex_path = os.path.join(DATA_DIR, complex_id)
complex_pdb_files = [file_ for file_ in os.listdir(complex_path) if file_.endswith('.pdb')]
complex_summary = apo_holo_summary_df[apo_holo_summary_df['file_name'].isin(complex_pdb_files)]
comparison_structures = complex_summary.query("structure_type == 'tcr' or state == 'holo'")
comparisons = pd.merge(comparison_structures, comparison_structures, how='cross')
comparisons['comparison'] = comparisons.apply(lambda row: '-'.join(sorted([row.file_name_x, row.file_name_y])),
axis='columns')
comparisons = comparisons.drop_duplicates('comparison')
comparisons = comparisons.drop('comparison', axis='columns')
comparisons = comparisons.query('file_name_x != file_name_y')
for _, comparison in comparisons.iterrows():
tcrs = []
for suffix in '_x', '_y':
with open(os.path.join(complex_path, comparison['file_name' + suffix]), 'r') as fh:
structure_df = parse_pdb_to_pandas(fh.read())
chains = comparison.filter(like='chain').filter(regex=f'{suffix}$').replace({np.nan: None}).tolist()
structure_df = annotate_tcr_pmhc_df(structure_df, *chains)
structure_df['backbone'] = structure_df['atom_name'].map(
lambda atom_name: (atom_name == 'N' or atom_name == 'CA' or atom_name == 'C' or atom_name == 'O')
)
tcr_df = structure_df.query("chain_type in ['alpha_chain', 'beta_chain']").copy().reset_index()
tcrs.append(tcr_df)
tcr_x, tcr_y = tcrs
# Compute C.O.Ms and coordinate vectors
calculations = defaultdict(dict)
for label, tcr_df in (('x', tcr_x), ('y', tcr_y)):
for chain_type in ('alpha_chain', 'beta_chain'):
anchors_ca = pd.DataFrame()
for cdr in (1, 2, 3):
cdr_df = tcr_df.query('chain_type == @chain_type and cdr == @cdr')
for cdr_anchor in find_anchors(cdr_df, tcr_df):
anchors_ca = pd.concat([anchors_ca, cdr_anchor.query("atom_name == 'CA'")])
fw_ca_df = tcr_df.query("cdr.isnull() and residue_seq_id <= 128 and chain_type == @chain_type and atom_name == 'CA'")
anchors_com = np.average(get_coords(anchors_ca), axis=0)
fw_com = np.average(get_coords(fw_ca_df), axis=0)
tcr_chain_direction_vec = fw_com - anchors_com
calculations[label][chain_type] = {}
calculations[label][chain_type]['anchors_com'] = anchors_com
calculations[label][chain_type]['fw_com'] = fw_com
calculations[label][chain_type]['direction_vector'] = tcr_chain_direction_vec
# Calculate differences
chain_anchor_com = []
chain_fw_com = []
for chain_type in ('alpha_chain', 'beta_chain'):
chain_anchor_com.append(get_distance(calculations['x'][chain_type]['anchors_com'],
calculations['y'][chain_type]['anchors_com']))
chain_fw_com.append(get_distance(calculations['x'][chain_type]['fw_com'],
calculations['y'][chain_type]['fw_com']))
angle_x = calculate_angle(calculations['x']['alpha_chain']['direction_vector'],
calculations['x']['beta_chain']['direction_vector'])
angle_y = calculate_angle(calculations['y']['alpha_chain']['direction_vector'],
calculations['y']['beta_chain']['direction_vector'])
angle_diff = angle_y - angle_x
# Collect Data
complex_ids.append(complex_id)
structure_x_paths.append(comparison['file_name_x'])
structure_y_paths.append(comparison['file_name_y'])
alpha_anchor_com_diffs.append(chain_anchor_com[0])
beta_anchor_com_diffs.append(chain_anchor_com[1])
alpha_fw_com_diffs.append(chain_fw_com[0])
beta_fw_com_diffs.append(chain_fw_com[1])
chain_angle_diffs.append(angle_diff)
results = pd.DataFrame({
'complex_id': complex_ids,
'structure_x_path': structure_x_paths,
'structure_y_path': structure_y_paths,
'alpha_anchor_com_diff': alpha_anchor_com_diffs,
'beta_anchor_com_diff': beta_anchor_com_diffs,
'alpha_fw_com_diff': alpha_fw_com_diffs,
'beta_fw_com_diff': beta_fw_com_diffs,
'chain_angle_diff': chain_angle_diffs,
})
results
[6]:
| complex_id | structure_x_path | structure_y_path | alpha_anchor_com_diff | beta_anchor_com_diff | alpha_fw_com_diff | beta_fw_com_diff | chain_angle_diff | |
|---|---|---|---|---|---|---|---|---|
| 0 | 3qdg_D-E-C-A-B_tcr_pmhc | 3qdg_D-E-C-A-B_tcr_pmhc.pdb | 3qeu_A-B_tcr.pdb | 0.645120 | 0.457327 | 0.219777 | 0.166987 | -0.033421 |
| 1 | 3qdg_D-E-C-A-B_tcr_pmhc | 3qdg_D-E-C-A-B_tcr_pmhc.pdb | 3qeu_D-E_tcr.pdb | 0.527090 | 0.340932 | 0.164026 | 0.213055 | -0.040502 |
| 2 | 3qdg_D-E-C-A-B_tcr_pmhc | 3qeu_A-B_tcr.pdb | 3qeu_D-E_tcr.pdb | 0.682250 | 0.284114 | 0.334339 | 0.064055 | -0.007081 |
| 3 | 5c0a_D-E-C-A-B_tcr_pmhc | 3utp_D-E_tcr.pdb | 3utp_K-L_tcr.pdb | 0.304534 | 0.602875 | 0.290645 | 0.338509 | 0.041175 |
| 4 | 5c0a_D-E-C-A-B_tcr_pmhc | 3utp_D-E_tcr.pdb | 5c0a_D-E-C-A-B_tcr_pmhc.pdb | 0.292352 | 0.514781 | 0.334429 | 0.270019 | 0.032863 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 131 | 5nme_D-E-C-A-B_tcr_pmhc | 5nmd_C-D_tcr.pdb | 5nme_D-E-C-A-B_tcr_pmhc.pdb | 0.211336 | 0.712433 | 0.113214 | 0.130459 | 0.030224 |
| 132 | 5hhm_D-E-C-A-B_tcr_pmhc | 2vlm_D-E_tcr.pdb | 5hhm_D-E-C-A-B_tcr_pmhc.pdb | 0.886794 | 0.410827 | 0.467935 | 0.381336 | -0.115053 |
| 133 | 3kpr_D-E-C-A-B_tcr_pmhc | 1kgc_D-E_tcr.pdb | 3kpr_D-E-C-A-B_tcr_pmhc.pdb | 0.919506 | 0.270885 | 0.383752 | 0.151581 | -0.026228 |
| 134 | 1oga_D-E-C-A-B_tcr_pmhc | 1oga_D-E-C-A-B_tcr_pmhc.pdb | 2vlm_D-E_tcr.pdb | 0.944931 | 0.542110 | 0.291887 | 0.336729 | 0.107774 |
| 135 | 7rtr_D-E-C-A-B_tcr_pmhc | 7n1d_A-B_tcr.pdb | 7rtr_D-E-C-A-B_tcr_pmhc.pdb | 0.642809 | 0.188609 | 0.388833 | 0.291080 | 0.001583 |
136 rows × 8 columns
[7]:
apo_holo_summary_df['ids'] = apo_holo_summary_df['file_name'].str.replace('.pdb', '')
/var/scratch/bmcmaste/2178229/ipykernel_2172418/2116153506.py:1: FutureWarning: The default value of regex will change from True to False in a future version.
apo_holo_summary_df['ids'] = apo_holo_summary_df['file_name'].str.replace('.pdb', '')
[8]:
results = results.merge(apo_holo_summary_df[['ids', 'cdr_sequences_collated', 'mhc_slug', 'peptide_sequence']],
how='left',
left_on='complex_id', right_on='ids')
[9]:
results = results.merge(
apo_holo_summary_df[['file_name', 'structure_type', 'state']],
how='left',
left_on='structure_x_path', right_on='file_name',
).merge(
apo_holo_summary_df[['file_name', 'structure_type', 'state']],
how='left',
left_on='structure_y_path', right_on='file_name',
)
[10]:
results['composite_name'] = results.apply(lambda row: '-'.join(sorted([row.structure_x_path, row.structure_y_path])), axis='columns')
results = results.drop_duplicates('composite_name')
[11]:
apo_holo_results = results.query('state_x != state_y')
[12]:
apo_holo_results_norm = apo_holo_results.groupby('cdr_sequences_collated').agg({
'alpha_anchor_com_diff': 'mean',
'beta_anchor_com_diff': 'mean',
'alpha_fw_com_diff': 'mean',
'beta_fw_com_diff': 'mean',
'chain_angle_diff': 'mean',
})
[13]:
apo_holo_results_norm['chain_angle_diff_deg'] = apo_holo_results_norm['chain_angle_diff'].apply(np.degrees)
apo_holo_results_norm['chain_angle_diff_deg_mag'] = apo_holo_results_norm['chain_angle_diff_deg'].apply(np.abs)
[14]:
apo_holo_results_norm
[14]:
| alpha_anchor_com_diff | beta_anchor_com_diff | alpha_fw_com_diff | beta_fw_com_diff | chain_angle_diff | chain_angle_diff_deg | chain_angle_diff_deg_mag | |
|---|---|---|---|---|---|---|---|
| cdr_sequences_collated | |||||||
| ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGRGDQPQH | 0.112478 | 0.221407 | 0.131706 | 0.140118 | -0.005545 | -0.317697 | 0.317697 |
| DRGSQS-IYSNGD-ALTRGPGNQFY-SGHVS-FNYEAQ-ASSSPGGVSTEAF | 1.606143 | 0.370279 | 1.293890 | 0.279836 | -0.057826 | -3.313194 | 3.313194 |
| DRGSQS-IYSNGD-AVNFGGGKLI-MRHNA-SNTAGT-ASSLSFGTEAF | 0.626083 | 0.366441 | 0.250095 | 0.178326 | -0.001998 | -0.114483 | 0.114483 |
| DRGSQS-IYSNGD-AVNRDDKII-SEHNR-FQNEAQ-ASSPDIEQY | 0.624668 | 0.204869 | 0.302308 | 0.186851 | -0.011943 | -0.684280 | 0.684280 |
| DRGSQS-IYSNGD-AVRTNSGYALN-QGHDT-YYEEEE-ASSDTVSYEQY | 0.306039 | 0.615471 | 0.133071 | 0.205778 | 0.024796 | 1.420686 | 1.420686 |
| DRGSQS-IYSNGD-AVTTDSWGKLQ-MNHEY-SVGAGI-ASRPGLAGGRPEQY | 0.385751 | 0.657747 | 0.300314 | 0.374858 | 0.018630 | 1.067402 | 1.067402 |
| DRGSQS-IYSNGD-GTYNQGGKLI-MNHEY-SMNVEV-ASSGASHEQY | 1.099042 | 0.294167 | 0.235768 | 0.174727 | 0.103824 | 5.948649 | 5.948649 |
| DSAIYN-IQSSQRE-AQLNQAGTALI-MNHEY-SVGAGI-ASSYGTGINYGYT | 0.596772 | 0.032190 | 0.184380 | 0.213406 | -0.037547 | -2.151258 | 2.151258 |
| DSAIYN-IQSSQRE-AVRMDSSYKLI-SEHNR-FQNEAQ-ASSSWDTGELF | 0.264265 | 0.248560 | 0.103594 | 0.098554 | -0.009340 | -0.535154 | 0.535154 |
| DSAIYN-IQSSQRE-AVRPLLDGTYIPT-MNHEY-SVGAGT-ASSYLGNTGELF | 0.408007 | 0.394163 | 0.483043 | 0.506659 | 0.044708 | 2.561604 | 2.561604 |
| DSAIYN-IQSSQRE-AVRPTSGGSYIPT-MNHEY-SVGAGI-ASSYVGNTGELF | 0.524052 | 0.405684 | 0.393972 | 0.306927 | -0.011481 | -0.657791 | 0.657791 |
| FLGSQS-TYREGD-AVNDGGRLT-GTSNPN-WGPFG-AWSETGLGMGGWQ | 0.520622 | 0.385860 | 0.254620 | 0.288849 | -0.009649 | -0.552852 | 0.552852 |
| NIATNDY-GYKTK-LVGEILDNFNKFY-MDHEN-SYDVKM-ASSQRQEGDTQY | 0.570818 | 0.282032 | 0.423455 | 0.414862 | 0.023776 | 1.362237 | 1.362237 |
| NSAFDY-ILSVSNK-AASASFGDNSKLI-MSHET-SYDVDS-ASSLGHTEVF | 0.124907 | 0.468265 | 0.138290 | 0.401072 | 0.016898 | 0.968205 | 0.968205 |
| NSAFQY-TYSSGN-AMRGDSSYKLI-SGHDY-FNNNVP-ASSLWEKLAKNIQY | 0.269449 | 0.373169 | 0.259665 | 0.247023 | 0.027624 | 1.582753 | 1.582753 |
| NSASQS-VYSSG-VVQPGGYQKVT-MNHNS-SASEGT-ASSEGLWQVGDEQY | 0.194964 | 0.279496 | 0.157453 | 0.355964 | 0.016874 | 0.966819 | 0.966819 |
| NSASQS-VYSSG-VVRAGKLI-MNHEY-SVGEGT-ASGQGNFDIQY | 0.409381 | 0.249897 | 0.205705 | 0.286431 | -0.055766 | -3.195168 | 3.195168 |
| SVFSS-VVTGGEV-AGAGSQGNLI-LNHDA-SQIVND-ASSSRSSYEQY | 1.003777 | 0.509446 | 0.331727 | 0.334128 | 0.058476 | 3.350441 | 3.350441 |
| TISGNEY-GLKNN-IVWGGYQKVT-SEHNR-FQNEAQ-ASRYRDDSYNEQF | 1.032154 | 1.282186 | 0.435426 | 0.393399 | 0.066787 | 3.826598 | 3.826598 |
| TISGTDY-GLTSN-ILPLAGGTSYGKLT-SGHVS-FQNEAQ-ASSLGQAYEQY | 0.713302 | 0.195773 | 0.230770 | 0.110227 | -0.002818 | -0.161464 | 0.161464 |
| YSATPY-YYSGDPVV-AVSGFASALT-NNHNN-SYGAGS-ASGGGGTLY | 0.304694 | 0.460448 | 0.099579 | 0.079669 | 0.006357 | 0.364241 | 0.364241 |
| YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGAGGADTQY | 0.796027 | 0.388025 | 0.043012 | 0.054271 | 0.112168 | 6.426748 | 6.426748 |
Visualizing Results
Changes in Framework (Fw) region and Loop Anchor Centre of Masses (COM)
[15]:
coms = apo_holo_results_norm.melt(value_vars=['alpha_anchor_com_diff', 'beta_anchor_com_diff', 'alpha_fw_com_diff', 'beta_fw_com_diff'],
var_name='location',
value_name='com_diff')
coms['chain_type'] = coms['location'].map(lambda location: location.split('_')[0])
coms['location'] = coms['location'].map(lambda location: location.split('_')[1])
[16]:
sns.boxplot(coms, y='com_diff', x='location', hue='chain_type')
[16]:
<AxesSubplot: xlabel='location', ylabel='com_diff'>
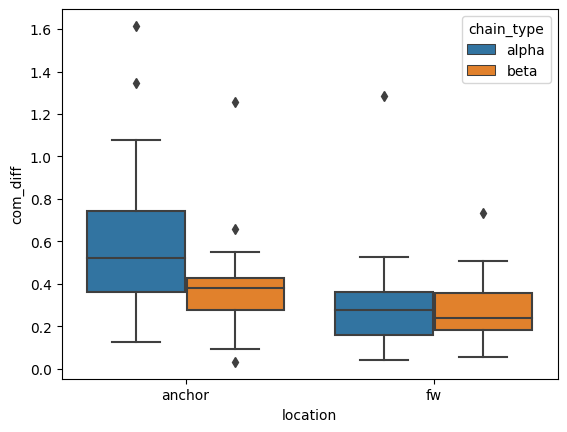
There is slight movement between the COMs but not very much.
Changes in angle between \(\alpha\)- and \(\beta\)- chain
[17]:
sns.boxplot(apo_holo_results_norm, y='chain_angle_diff')
apo_holo_results_norm['chain_angle_diff'].describe()
[17]:
count 22.000000
mean 0.014409
std 0.043607
min -0.057826
25% -0.009572
50% 0.011616
75% 0.026917
max 0.112168
Name: chain_angle_diff, dtype: float64
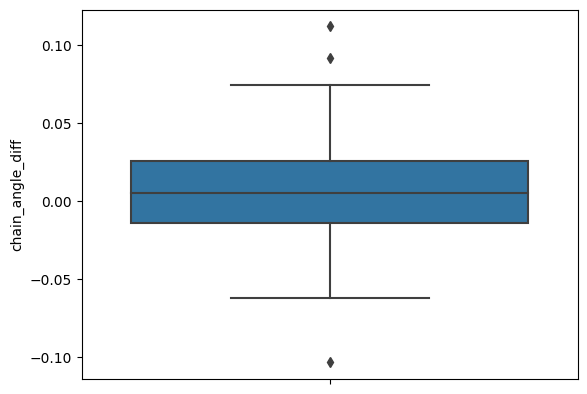
[18]:
sns.boxplot(apo_holo_results_norm, y='chain_angle_diff_deg')
apo_holo_results_norm['chain_angle_diff_deg'].describe()
[18]:
count 22.000000
mean 0.825593
std 2.498524
min -3.313194
25% -0.548427
50% 0.665530
75% 1.542237
max 6.426748
Name: chain_angle_diff_deg, dtype: float64
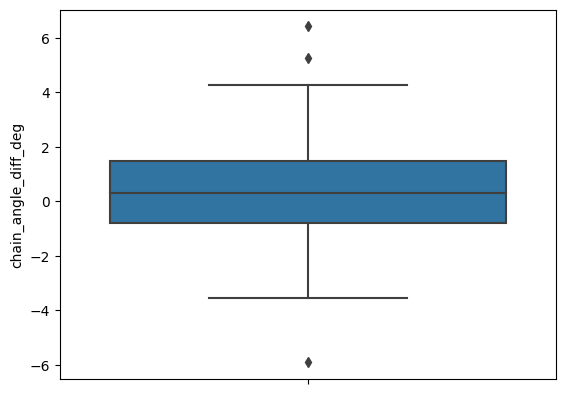
[19]:
sns.boxplot(apo_holo_results_norm, y='chain_angle_diff_deg_mag')
apo_holo_results_norm['chain_angle_diff_deg_mag'].describe()
[19]:
count 22.000000
mean 1.887715
std 1.795418
min 0.114483
25% 0.579086
50% 1.214819
75% 3.036777
max 6.426748
Name: chain_angle_diff_deg_mag, dtype: float64
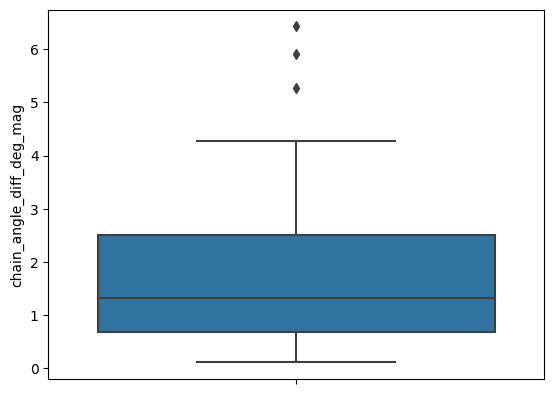
From these plots, it does not seem that their is a lot of angular change between domains.
Conclusion
Based on the results of this analysis, the TCR Fw region remains fairly static between apo and holo.