Comparing apo and holo CDR loop clustering
Introduction
In this notebook, we set out to assess whether the CDR loops of TCRs stay in there canonical forms between apo and holo states or if they disrupt them.
[1]:
import itertools
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
[2]:
df = pd.read_csv('../data/processed/stcrdab_clusters.csv')
df
[2]:
| name | cluster | chain_type | cdr | sequence | cluster_type | |
|---|---|---|---|---|---|---|
| 0 | 7zt2_DE | 12 | alpha_chain | 1 | TSGFNG | pseudo |
| 1 | 7zt3_DE | 12 | alpha_chain | 1 | TSGFNG | pseudo |
| 2 | 7zt4_DE | 12 | alpha_chain | 1 | TSGFNG | pseudo |
| 3 | 7zt5_DE | 12 | alpha_chain | 1 | TSGFNG | pseudo |
| 4 | 7zt7_DE | 12 | alpha_chain | 1 | TSGFNG | pseudo |
| ... | ... | ... | ... | ... | ... | ... |
| 4807 | 6miv_CD | 22 | beta_chain | 3 | ASGDEGYTQY | canonical |
| 4808 | 3rtq_CD | 22 | beta_chain | 3 | ASGDEGYTQY | canonical |
| 4809 | 3dxa_NO | noise | beta_chain | 3 | ASRYRDDSYNEQF | NaN |
| 4810 | 1d9k_AB | noise | beta_chain | 3 | ASGGQGRAEQF | NaN |
| 4811 | 4gg6_GH | noise | beta_chain | 3 | ASSVAVSAGTYEQY | NaN |
4812 rows × 6 columns
Compare apo and holo forms of loops
[3]:
apo_holo_summary_df = pd.read_csv('../data/processed/apo-holo-tcr-pmhc-class-I/apo_holo_summary.csv')
apo_holo_summary_df
[3]:
| file_name | pdb_id | structure_type | state | alpha_chain | beta_chain | antigen_chain | mhc_chain1 | mhc_chain2 | cdr_sequences_collated | peptide_sequence | mhc_slug | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1ao7_D-E-C-A-B_tcr_pmhc.pdb | 1ao7 | tcr_pmhc | holo | D | E | C | A | B | DRGSQS-IYSNGD-AVTTDSWGKLQ-MNHEY-SVGAGI-ASRPGLA... | LLFGYPVYV | hla_a_02_01 |
| 1 | 1b0g_C-A-B_pmhc.pdb | 1b0g | pmhc | apo | NaN | NaN | C | A | B | NaN | ALWGFFPVL | hla_a_02_01 |
| 2 | 1b0g_F-D-E_pmhc.pdb | 1b0g | pmhc | apo | NaN | NaN | F | D | E | NaN | ALWGFFPVL | hla_a_02_01 |
| 3 | 1bd2_D-E-C-A-B_tcr_pmhc.pdb | 1bd2 | tcr_pmhc | holo | D | E | C | A | B | NSMFDY-ISSIKDK-AAMEGAQKLV-MNHEY-SVGAGI-ASSYPGG... | LLFGYPVYV | hla_a_02_01 |
| 4 | 1bii_P-A-B_pmhc.pdb | 1bii | pmhc | apo | NaN | NaN | P | A | B | NaN | RGPGRAFVTI | h2_dd |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 386 | 7rtd_C-A-B_pmhc.pdb | 7rtd | pmhc | apo | NaN | NaN | C | A | B | NaN | YLQPRTFLL | hla_a_02_01 |
| 387 | 7rtr_D-E-C-A-B_tcr_pmhc.pdb | 7rtr | tcr_pmhc | holo | D | E | C | A | B | DRGSQS-IYSNGD-AVNRDDKII-SEHNR-FQNEAQ-ASSPDIEQY | YLQPRTFLL | hla_a_02_01 |
| 388 | 8gvb_A-B-P-H-L_tcr_pmhc.pdb | 8gvb | tcr_pmhc | holo | A | B | P | H | L | YGATPY-YFSGDTLV-AVGFTGGGNKLT-SEHNR-FQNEAQ-ASSD... | RYPLTFGW | hla_a_24_02 |
| 389 | 8gvg_A-B-P-H-L_tcr_pmhc.pdb | 8gvg | tcr_pmhc | holo | A | B | P | H | L | YGATPY-YFSGDTLV-AVGFTGGGNKLT-SEHNR-FQNEAQ-ASSD... | RFPLTFGW | hla_a_24_02 |
| 390 | 8gvi_A-B-P-H-L_tcr_pmhc.pdb | 8gvi | tcr_pmhc | holo | A | B | P | H | L | YGATPY-YFSGDTLV-AVVFTGGGNKLT-SEHNR-FQNEAQ-ASSL... | RYPLTFGW | hla_a_24_02 |
391 rows × 12 columns
[4]:
df[['pdb_id', 'chains']] = df['name'].str.split('_').apply(pd.Series)
df[['alpha_chain', 'beta_chain']] = df['chains'].apply(list).apply(pd.Series)
[5]:
apo_holo_clusters = df.merge(apo_holo_summary_df,
how='inner',
on=['pdb_id', 'alpha_chain', 'beta_chain'])
apo_holo_clusters
[5]:
| name | cluster | chain_type | cdr | sequence | cluster_type | pdb_id | chains | alpha_chain | beta_chain | file_name | structure_type | state | antigen_chain | mhc_chain1 | mhc_chain2 | cdr_sequences_collated | peptide_sequence | mhc_slug | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2ak4_DE | 4 | alpha_chain | 1 | TRDTTYY | canonical | 2ak4 | DE | D | E | 2ak4_D-E-C-A-B_tcr_pmhc.pdb | tcr_pmhc | holo | C | A | B | TRDTTYY-RNSFDEQN-ALSGFYNTDKLI-MNHNS-SASEGT-ASP... | LPEPLPQGQLTAY | hla_b_35_08 |
| 1 | 2ak4_DE | 6 | beta_chain | 1 | MNHNS | canonical | 2ak4 | DE | D | E | 2ak4_D-E-C-A-B_tcr_pmhc.pdb | tcr_pmhc | holo | C | A | B | TRDTTYY-RNSFDEQN-ALSGFYNTDKLI-MNHNS-SASEGT-ASP... | LPEPLPQGQLTAY | hla_b_35_08 |
| 2 | 2ak4_DE | 6 | alpha_chain | 2 | RNSFDEQN | pseudo | 2ak4 | DE | D | E | 2ak4_D-E-C-A-B_tcr_pmhc.pdb | tcr_pmhc | holo | C | A | B | TRDTTYY-RNSFDEQN-ALSGFYNTDKLI-MNHNS-SASEGT-ASP... | LPEPLPQGQLTAY | hla_b_35_08 |
| 3 | 2ak4_DE | noise | beta_chain | 2 | SASEGT | NaN | 2ak4 | DE | D | E | 2ak4_D-E-C-A-B_tcr_pmhc.pdb | tcr_pmhc | holo | C | A | B | TRDTTYY-RNSFDEQN-ALSGFYNTDKLI-MNHNS-SASEGT-ASP... | LPEPLPQGQLTAY | hla_b_35_08 |
| 4 | 2ak4_DE | 12 | alpha_chain | 3 | ALSGFYNTDKLI | pseudo | 2ak4 | DE | D | E | 2ak4_D-E-C-A-B_tcr_pmhc.pdb | tcr_pmhc | holo | C | A | B | TRDTTYY-RNSFDEQN-ALSGFYNTDKLI-MNHNS-SASEGT-ASP... | LPEPLPQGQLTAY | hla_b_35_08 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1105 | 4jff_DE | 0 | beta_chain | 1 | GTSNPN | pseudo | 4jff | DE | D | E | 4jff_D-E-C-A-B_tcr_pmhc.pdb | tcr_pmhc | holo | C | A | B | FLGSQS-TYREGD-AVNDGGRLT-GTSNPN-WGPFG-AWSETGLGM... | ELAGIGILTV | hla_a_02_01 |
| 1106 | 4jff_DE | 17 | alpha_chain | 2 | TYREGD | canonical | 4jff | DE | D | E | 4jff_D-E-C-A-B_tcr_pmhc.pdb | tcr_pmhc | holo | C | A | B | FLGSQS-TYREGD-AVNDGGRLT-GTSNPN-WGPFG-AWSETGLGM... | ELAGIGILTV | hla_a_02_01 |
| 1107 | 4jff_DE | noise | beta_chain | 2 | WGPFG | NaN | 4jff | DE | D | E | 4jff_D-E-C-A-B_tcr_pmhc.pdb | tcr_pmhc | holo | C | A | B | FLGSQS-TYREGD-AVNDGGRLT-GTSNPN-WGPFG-AWSETGLGM... | ELAGIGILTV | hla_a_02_01 |
| 1108 | 4jff_DE | 26 | alpha_chain | 3 | AVNDGGRLT | canonical | 4jff | DE | D | E | 4jff_D-E-C-A-B_tcr_pmhc.pdb | tcr_pmhc | holo | C | A | B | FLGSQS-TYREGD-AVNDGGRLT-GTSNPN-WGPFG-AWSETGLGM... | ELAGIGILTV | hla_a_02_01 |
| 1109 | 4jff_DE | noise | beta_chain | 3 | AWSETGLGMGGWQ | NaN | 4jff | DE | D | E | 4jff_D-E-C-A-B_tcr_pmhc.pdb | tcr_pmhc | holo | C | A | B | FLGSQS-TYREGD-AVNDGGRLT-GTSNPN-WGPFG-AWSETGLGM... | ELAGIGILTV | hla_a_02_01 |
1110 rows × 19 columns
[6]:
def collate_clusters(group):
apo_clusters = sorted(group.query("state == 'apo'")['cluster'].tolist())
holo_clusters = sorted(group.query("state == 'holo'")['cluster'].tolist())
return pd.Series({'apo_clusters': apo_clusters, 'holo_clusters': holo_clusters})
apo_holo_clusters_agg = apo_holo_clusters.groupby(['cdr_sequences_collated',
'chain_type',
'cdr']).apply(collate_clusters).reset_index()
apo_holo_clusters_agg
[6]:
| cdr_sequences_collated | chain_type | cdr | apo_clusters | holo_clusters | |
|---|---|---|---|---|---|
| 0 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 1 | [13] | [noise] |
| 1 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 2 | [noise] | [noise] |
| 2 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 3 | [noise] | [noise] |
| 3 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | beta_chain | 1 | [4] | [4] |
| 4 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | beta_chain | 2 | [2] | [2] |
| ... | ... | ... | ... | ... | ... |
| 511 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | alpha_chain | 2 | [noise] | [noise] |
| 512 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | alpha_chain | 3 | [noise] | [2] |
| 513 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 1 | [4] | [4] |
| 514 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 2 | [2] | [2] |
| 515 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 3 | [noise] | [noise] |
516 rows × 5 columns
[7]:
cluster_types = (df[['chain_type', 'cdr', 'cluster', 'cluster_type']].drop_duplicates()
.set_index(['chain_type', 'cdr', 'cluster'])
['cluster_type'])
[8]:
def classify_movement(chain_type: str,
cdr: str,
apo_clusters: list[str],
holo_clusters: list[str],
cluster_types: pd.Series) -> pd.Series:
types = {
'Noise': 0,
'Noise to Canonical Cluster': 0,
'Noise to Pseudo Cluster': 0,
'Canonical Cluster to Noise': 0,
'Pseudo Cluster to Noise': 0,
'Canonical Cluster Shift': 0,
'Pseudo Cluster Shift': 0,
'Canonical to Pseudo': 0,
'Pseudo to Canonical': 0,
'Canonical Cluster Same': 0,
'Pseudo Cluster Same': 0,
}
for apo_clust, holo_clust in itertools.product(apo_clusters, holo_clusters):
if apo_clust == 'noise' and holo_clust == 'noise':
types['Noise'] += 1
elif apo_clust == 'noise' and holo_clust != 'noise':
if cluster_types.loc[chain_type, cdr, holo_clust] == 'canonical':
types['Noise to Canonical Cluster'] += 1
else:
types['Noise to Pseudo Cluster'] += 1
elif apo_clust != 'noise' and holo_clust == 'noise':
if cluster_types.loc[chain_type, cdr, apo_clust] == 'canonical':
types['Canonical Cluster to Noise'] += 1
else:
types['Pseudo Cluster to Noise'] += 1
elif apo_clust != holo_clust:
if (cluster_types.loc[chain_type, cdr, apo_clust]
== cluster_types.loc[chain_type, cdr, holo_clust]
== 'canonical'):
types['Canonical Cluster Shift'] += 1
elif (cluster_types.loc[chain_type, cdr, apo_clust] == 'canonical'
and cluster_types.loc[chain_type, cdr, holo_clust] == 'pseudo'):
types['Canonical to Pseudo'] += 1
else:
types['Pseudo to Canonical'] += 1
elif apo_clust == holo_clust:
if cluster_types.loc[chain_type, cdr, apo_clust] == 'canonical':
types['Canonical Cluster Same'] += 1
else:
types['Pseudo Cluster Same'] += 1
return pd.Series(types)
apo_holo_clusters_agg = apo_holo_clusters_agg.join(apo_holo_clusters_agg.apply(
lambda row: classify_movement(row.chain_type, row.cdr, row.apo_clusters, row.holo_clusters, cluster_types), axis=1
))
# Wide to long
apo_holo_clusters_agg = apo_holo_clusters_agg.melt(id_vars=['cdr_sequences_collated', 'chain_type', 'cdr'],
value_vars=['Noise',
'Noise to Canonical Cluster',
'Noise to Pseudo Cluster',
'Canonical Cluster to Noise',
'Pseudo Cluster to Noise',
'Canonical Cluster Shift',
'Pseudo Cluster Shift',
'Canonical to Pseudo',
'Pseudo to Canonical',
'Canonical Cluster Same',
'Pseudo Cluster Same'],
var_name='types', value_name='count')
# Normalize for differeing group sizes
apo_holo_clusters_agg = apo_holo_clusters_agg.groupby(
['cdr_sequences_collated', 'chain_type', 'cdr', 'types'],
)['count'].median().reset_index()
apo_holo_clusters_agg
[8]:
| cdr_sequences_collated | chain_type | cdr | types | count | |
|---|---|---|---|---|---|
| 0 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 1 | Canonical Cluster Same | 0.0 |
| 1 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 1 | Canonical Cluster Shift | 0.0 |
| 2 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 1 | Canonical Cluster to Noise | 1.0 |
| 3 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 1 | Canonical to Pseudo | 0.0 |
| 4 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 1 | Noise | 0.0 |
| ... | ... | ... | ... | ... | ... |
| 5671 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 3 | Noise to Pseudo Cluster | 0.0 |
| 5672 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 3 | Pseudo Cluster Same | 0.0 |
| 5673 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 3 | Pseudo Cluster Shift | 0.0 |
| 5674 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 3 | Pseudo Cluster to Noise | 0.0 |
| 5675 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 3 | Pseudo to Canonical | 0.0 |
5676 rows × 5 columns
[9]:
apo_holo_clusters_agg
[9]:
| cdr_sequences_collated | chain_type | cdr | types | count | |
|---|---|---|---|---|---|
| 0 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 1 | Canonical Cluster Same | 0.0 |
| 1 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 1 | Canonical Cluster Shift | 0.0 |
| 2 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 1 | Canonical Cluster to Noise | 1.0 |
| 3 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 1 | Canonical to Pseudo | 0.0 |
| 4 | ATGYPS-ATKADDK-ALSDPVNDMR-SGHAT-FQNNGV-ASSLRGR... | alpha_chain | 1 | Noise | 0.0 |
| ... | ... | ... | ... | ... | ... |
| 5671 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 3 | Noise to Pseudo Cluster | 0.0 |
| 5672 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 3 | Pseudo Cluster Same | 0.0 |
| 5673 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 3 | Pseudo Cluster Shift | 0.0 |
| 5674 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 3 | Pseudo Cluster to Noise | 0.0 |
| 5675 | YSGSPE-HISR-ALSGFNNAGNMLT-SGHAT-FQNNGV-ASSLGGA... | beta_chain | 3 | Pseudo to Canonical | 0.0 |
5676 rows × 5 columns
[10]:
g = sns.catplot(apo_holo_clusters_agg.sort_values(['chain_type', 'cdr', 'types']),
row='chain_type', col='cdr',
x='types', y='count',
kind='bar')
for ax in g.axes.flat:
labels = ax.get_xticklabels()
plt.setp(labels, rotation=90)
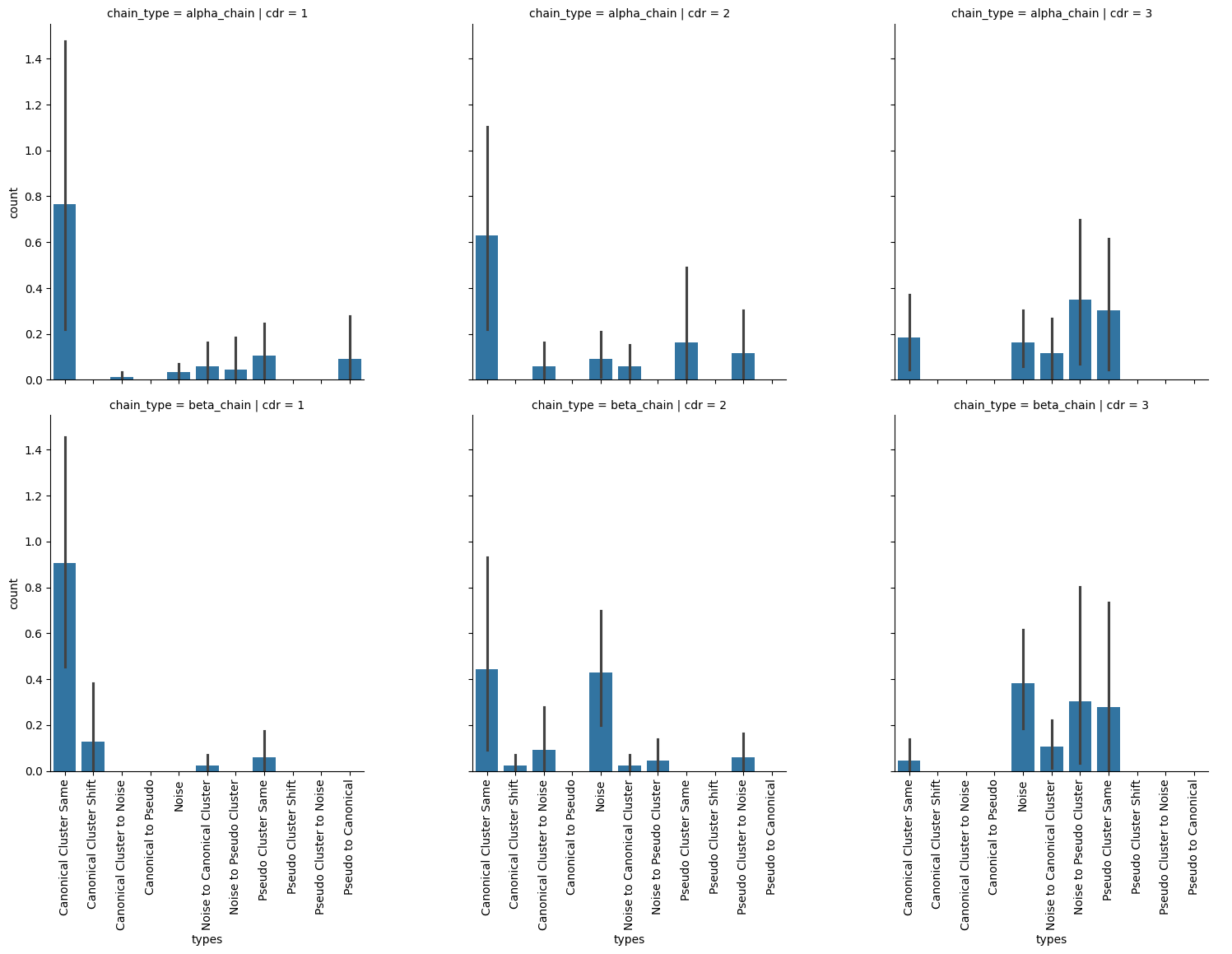
[11]:
cluster_shift_counts = apo_holo_clusters_agg.groupby(['chain_type', 'cdr', 'types'])['count'].sum().astype(int)
cluster_shift_counts[cluster_shift_counts > 0]
[11]:
chain_type cdr types
alpha_chain 1 Canonical Cluster Same 66
Canonical Cluster to Noise 1
Noise 3
Noise to Canonical Cluster 5
Noise to Pseudo Cluster 4
Pseudo Cluster Same 9
Pseudo to Canonical 8
2 Canonical Cluster Same 54
Canonical Cluster to Noise 5
Noise 8
Noise to Canonical Cluster 5
Pseudo Cluster Same 14
Pseudo Cluster to Noise 10
3 Canonical Cluster Same 16
Noise 14
Noise to Canonical Cluster 10
Noise to Pseudo Cluster 30
Pseudo Cluster Same 26
beta_chain 1 Canonical Cluster Same 78
Canonical Cluster Shift 11
Noise to Canonical Cluster 2
Pseudo Cluster Same 5
2 Canonical Cluster Same 38
Canonical Cluster Shift 2
Canonical Cluster to Noise 8
Noise 37
Noise to Canonical Cluster 2
Noise to Pseudo Cluster 4
Pseudo Cluster to Noise 5
3 Canonical Cluster Same 4
Noise 33
Noise to Canonical Cluster 9
Noise to Pseudo Cluster 26
Pseudo Cluster Same 24
Name: count, dtype: int64
[ ]:
export_cluster_shift_counts = cluster_shift_counts.to_frame().reset_index()
export_cluster_shift_counts = export_cluster_shift_counts.rename({'types': 'Movement Type'}, axis='columns')
export_cluster_shift_counts['CDR Type'] = export_cluster_shift_counts.apply(
lambda row: f"CDR{row.cdr}\\text{row.chain_type.split('_')[0]}{{}}",
axis='columns',
)
export_cluster_shift_counts = export_cluster_shift_counts.pivot(index='Movement Type',
columns='CDR Type',
values='count')
export_cluster_shift_counts = export_cluster_shift_counts.sort_values('Movement Type')
export_cluster_shift_counts = export_cluster_shift_counts.replace(0, '-')
export_cluster_shift_counts = export_cluster_shift_counts.sort_index()
export_cluster_shift_counts.columns.name = None
export_cluster_shift_counts = export_cluster_shift_counts.reset_index()
export_cluster_shift_counts
| Movement Type | CDR1\textalpha{} | CDR1\textbeta{} | CDR2\textalpha{} | CDR2\textbeta{} | CDR3\textalpha{} | CDR3\textbeta{} | |
|---|---|---|---|---|---|---|---|
| 0 | Canonical Cluster Same | 66 | 78 | 54 | 38 | 16 | 4 |
| 1 | Canonical Cluster Shift | - | 11 | - | 2 | - | - |
| 2 | Canonical Cluster to Noise | 1 | - | 5 | 8 | - | - |
| 3 | Canonical to Pseudo | - | - | - | - | - | - |
| 4 | Noise | 3 | - | 8 | 37 | 14 | 33 |
| 5 | Noise to Canonical Cluster | 5 | 2 | 5 | 2 | 10 | 9 |
| 6 | Noise to Pseudo Cluster | 4 | - | - | 4 | 30 | 26 |
| 7 | Pseudo Cluster Same | 9 | 5 | 14 | - | 26 | 24 |
| 8 | Pseudo Cluster Shift | - | - | - | - | - | - |
| 9 | Pseudo Cluster to Noise | - | - | 10 | 5 | - | - |
| 10 | Pseudo to Canonical | 8 | - | - | - | - | - |
Conclusion
The analysis here shows that for the most part, the CDR-1s and CDR-2s stay in their canonical forms between apo and holo states where as the CDR-3 loops have a variety of modes. As expected, it is more difficult to cluster the CDR-3 loops as they are more structurally diverse.