Visualising CDR loop clustering
Introduction
In this notebook, we visualise the clusters of loop conformations determined by the pair-wise distance comparison of all loops.
[1]:
import glob
import os
import subprocess
import tempfile
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import plotly.express as px
import seaborn as sns
import logomaker
from ipywidgets import Dropdown, VBox
from IPython.display import display, clear_output
from python_pdb.aligners import align_pandas_structure
from python_pdb.parsers import parse_pdb_to_pandas
from sklearn.manifold import TSNE
from tcr_pmhc_interface_analysis.processing import annotate_tcr_pmhc_df, find_anchors
from tcr_pmhc_interface_analysis.utils import get_coords
[2]:
DATA_DIR = '../data/interim/structure-pw-distances'
[3]:
with open(os.path.join(DATA_DIR, 'structure_names.txt'), 'r') as fh:
structure_names = [line.strip() for line in fh.readlines()]
[4]:
cdr_clusters = pd.read_csv('../data/processed/stcrdab_clusters.csv')
cdr_clusters
[4]:
| name | cluster | chain_type | cdr | sequence | cluster_type | |
|---|---|---|---|---|---|---|
| 0 | 7zt2_DE | 12 | alpha_chain | 1 | TSGFNG | pseudo |
| 1 | 7zt3_DE | 12 | alpha_chain | 1 | TSGFNG | pseudo |
| 2 | 7zt4_DE | 12 | alpha_chain | 1 | TSGFNG | pseudo |
| 3 | 7zt5_DE | 12 | alpha_chain | 1 | TSGFNG | pseudo |
| 4 | 7zt7_DE | 12 | alpha_chain | 1 | TSGFNG | pseudo |
| ... | ... | ... | ... | ... | ... | ... |
| 4807 | 6miv_CD | 22 | beta_chain | 3 | ASGDEGYTQY | canonical |
| 4808 | 3rtq_CD | 22 | beta_chain | 3 | ASGDEGYTQY | canonical |
| 4809 | 3dxa_NO | noise | beta_chain | 3 | ASRYRDDSYNEQF | NaN |
| 4810 | 1d9k_AB | noise | beta_chain | 3 | ASGGQGRAEQF | NaN |
| 4811 | 4gg6_GH | noise | beta_chain | 3 | ASSVAVSAGTYEQY | NaN |
4812 rows × 6 columns
[5]:
df = pd.DataFrame()
for path in glob.glob(os.path.join(DATA_DIR, '*_distance_matrix.txt*')):
name = os.path.basename(path).split('.')[0].replace('_distance_matrix', '')
cdr, chain = name.split('_')
cdr_distance_matrix = np.loadtxt(path)
cdr_tsne = TSNE(metric='precomputed', init='random', learning_rate='auto').fit_transform(cdr_distance_matrix)
cdr_df = pd.DataFrame({
'name': structure_names,
'tsne_1': cdr_tsne[:, 0],
'tsne_2': cdr_tsne[:, 1],
})
cdr_df['chain_type'] = chain + '_chain'
cdr_df['cdr'] = int(cdr.replace('cdr', ''))
df = pd.concat([df, cdr_df])
df
[5]:
| name | tsne_1 | tsne_2 | chain_type | cdr | |
|---|---|---|---|---|---|
| 0 | 7zt2_DE | -12.995802 | 14.408054 | beta_chain | 2 |
| 1 | 7zt3_DE | -12.007703 | 13.610337 | beta_chain | 2 |
| 2 | 7zt4_DE | -12.722438 | 14.317860 | beta_chain | 2 |
| 3 | 7zt5_DE | -12.896334 | 14.089248 | beta_chain | 2 |
| 4 | 7zt7_DE | -12.443920 | 14.690278 | beta_chain | 2 |
| ... | ... | ... | ... | ... | ... |
| 797 | 6miv_CD | -7.597044 | 46.012802 | alpha_chain | 3 |
| 798 | 3rtq_CD | -11.724574 | 50.600700 | alpha_chain | 3 |
| 799 | 3dxa_NO | -28.833708 | 18.615999 | alpha_chain | 3 |
| 800 | 1d9k_AB | -18.946213 | 13.313275 | alpha_chain | 3 |
| 801 | 4gg6_GH | -25.602039 | 18.878851 | alpha_chain | 3 |
4812 rows × 5 columns
[6]:
df = df.merge(cdr_clusters, how='inner', on=['name', 'chain_type', 'cdr'])
df
[6]:
| name | tsne_1 | tsne_2 | chain_type | cdr | cluster | sequence | cluster_type | |
|---|---|---|---|---|---|---|---|---|
| 0 | 7zt2_DE | -12.995802 | 14.408054 | beta_chain | 2 | 10 | SASEGT | pseudo |
| 1 | 7zt3_DE | -12.007703 | 13.610337 | beta_chain | 2 | 10 | SASEGT | pseudo |
| 2 | 7zt4_DE | -12.722438 | 14.317860 | beta_chain | 2 | 10 | SASEGT | pseudo |
| 3 | 7zt5_DE | -12.896334 | 14.089248 | beta_chain | 2 | 10 | SASEGT | pseudo |
| 4 | 7zt7_DE | -12.443920 | 14.690278 | beta_chain | 2 | 10 | SASEGT | pseudo |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4807 | 6miv_CD | -7.597044 | 46.012802 | alpha_chain | 3 | 13 | VVGDRGSALGRLH | canonical |
| 4808 | 3rtq_CD | -11.724574 | 50.600700 | alpha_chain | 3 | 13 | VVGDRGSALGRLH | canonical |
| 4809 | 3dxa_NO | -28.833708 | 18.615999 | alpha_chain | 3 | 6 | IVWGGYQKVT | canonical |
| 4810 | 1d9k_AB | -18.946213 | 13.313275 | alpha_chain | 3 | noise | AATGSFNKLT | NaN |
| 4811 | 4gg6_GH | -25.602039 | 18.878851 | alpha_chain | 3 | noise | ILRDGRGGADGLT | NaN |
4812 rows × 8 columns
[7]:
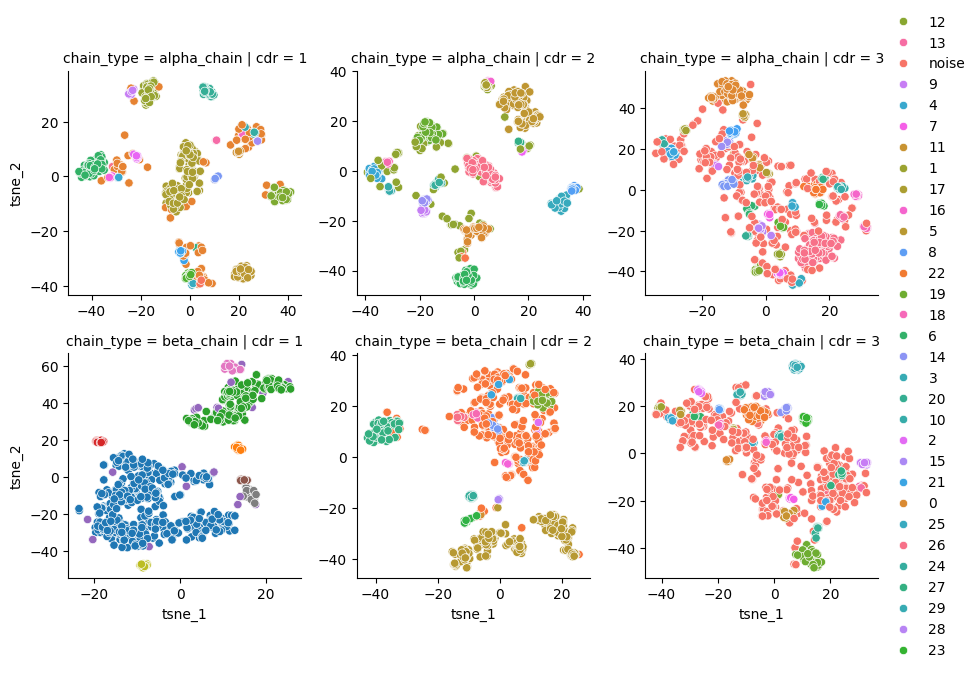
g = sns.FacetGrid(df.sort_values(['chain_type', 'cdr']), row='chain_type', col='cdr', sharex=False, sharey=False)
g.map(sns.scatterplot, 'tsne_1', 'tsne_2', 'cluster')
g.add_legend()
[7]:
<seaborn.axisgrid.FacetGrid at 0x7f251c37b460>
[8]:
df.query("cluster != 'noise'").groupby(['chain_type', 'cdr'])['cluster'].nunique()
[8]:
chain_type cdr
alpha_chain 1 23
2 18
3 30
beta_chain 1 8
2 16
3 28
Name: cluster, dtype: int64
Do the canonical clusters exist across different loop lengths?
[9]:
df['cdr_length'] = df['sequence'].map(len)
[10]:
df.query("cluster != 'noise'").groupby(['chain_type', 'cdr', 'cluster'])['cdr_length'].nunique().value_counts()
[10]:
1 107
2 14
3 2
Name: cdr_length, dtype: int64
Some clustering with different lengths!
Visualizing the canonical cluster structures and sequences
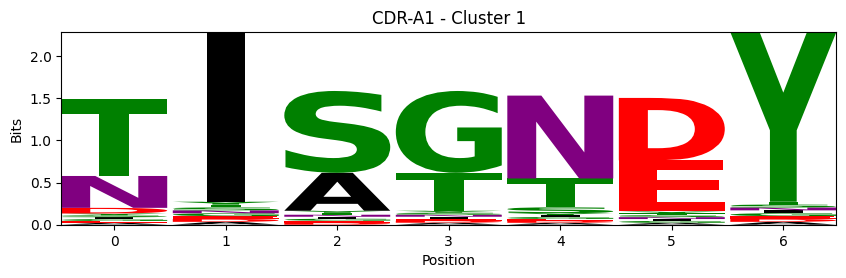
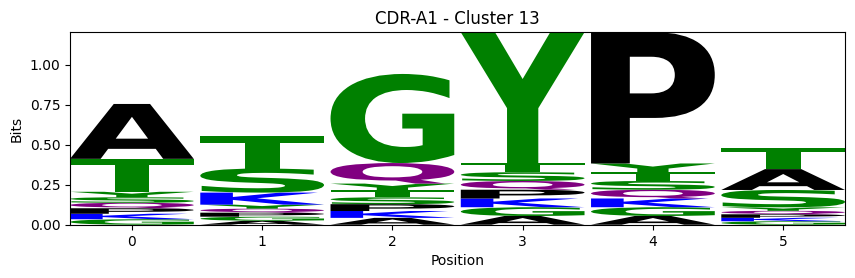
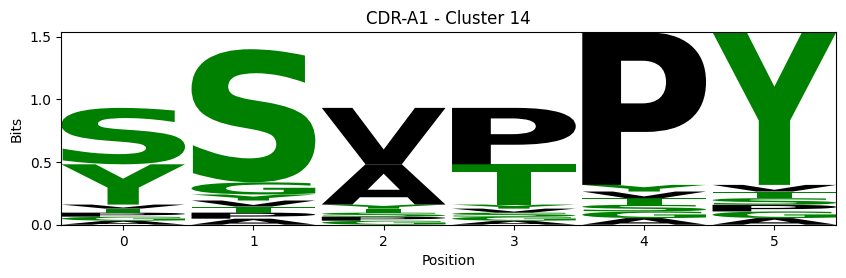
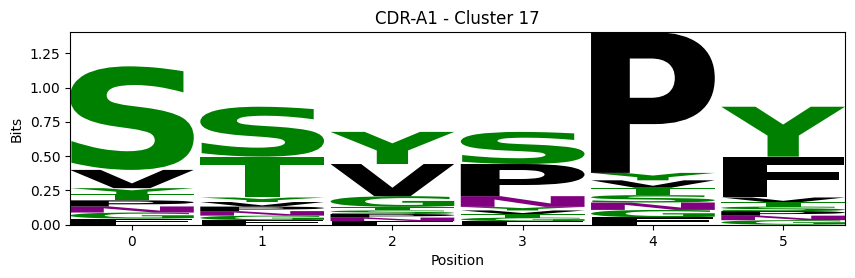
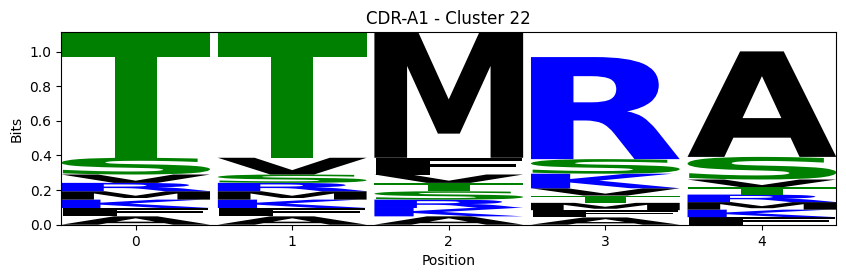
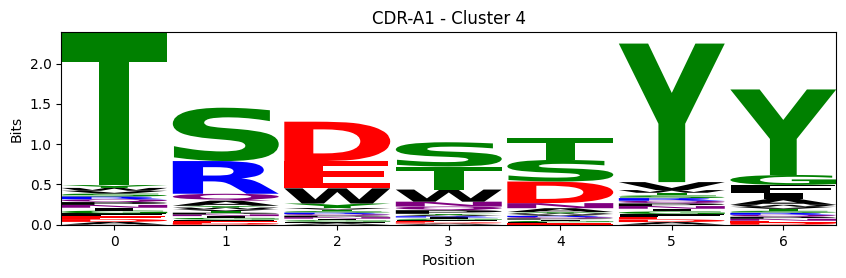
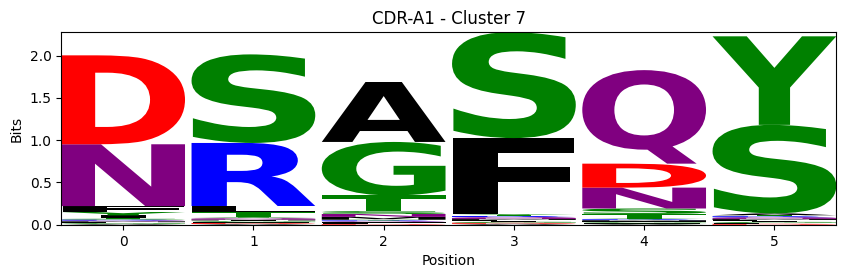
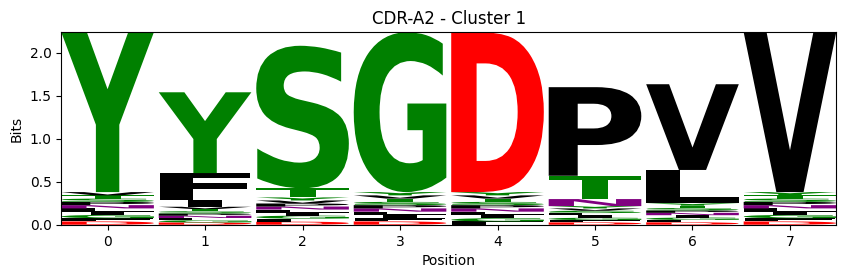
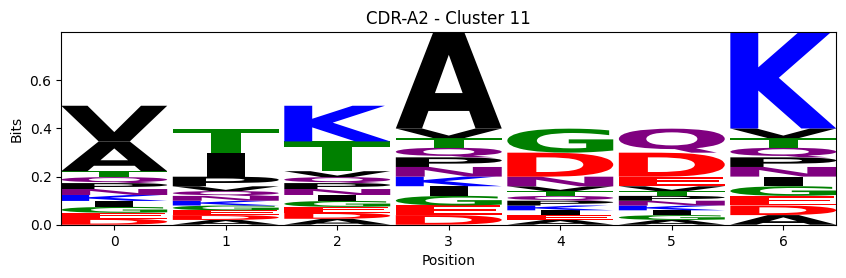
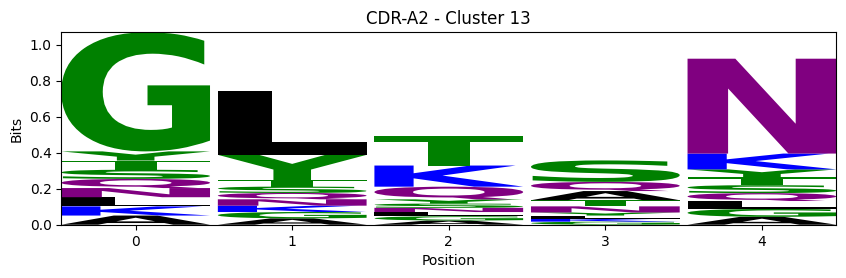
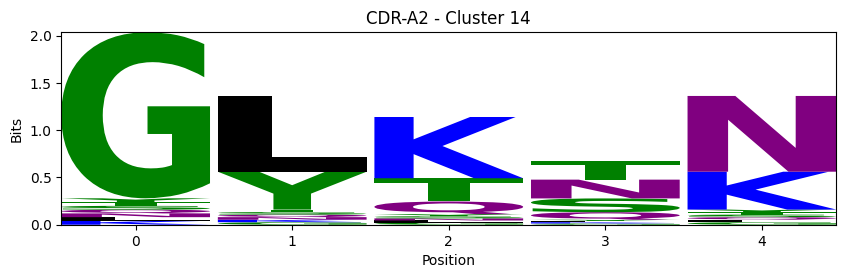
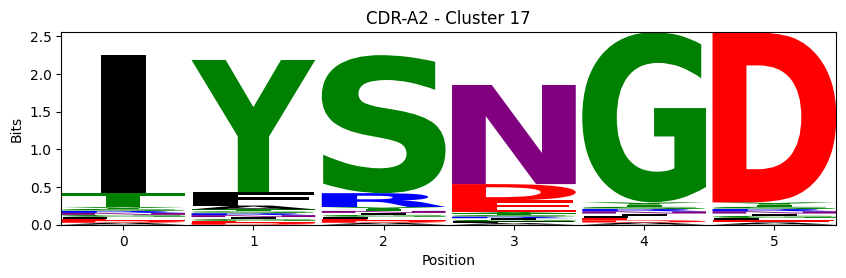
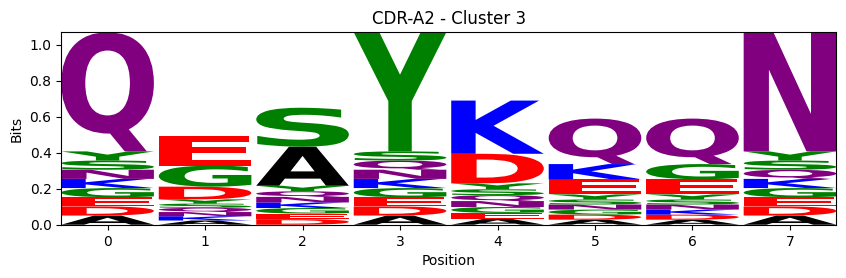
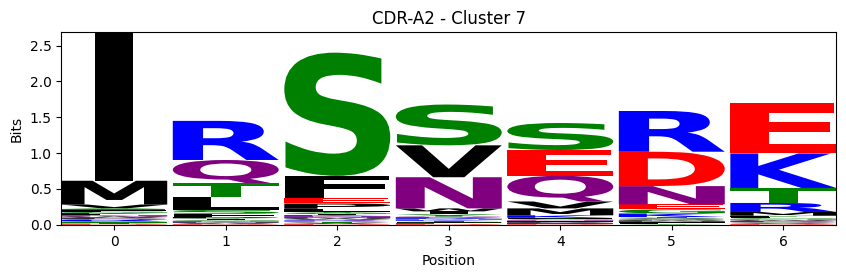
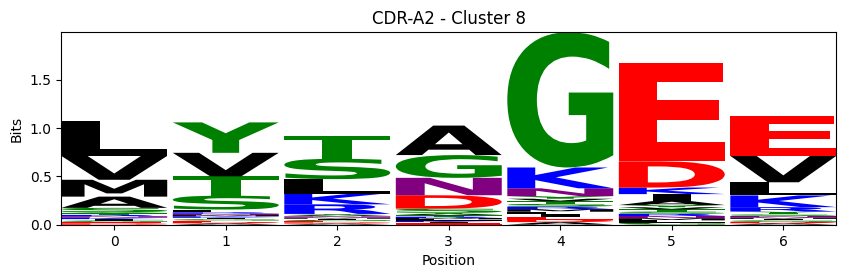
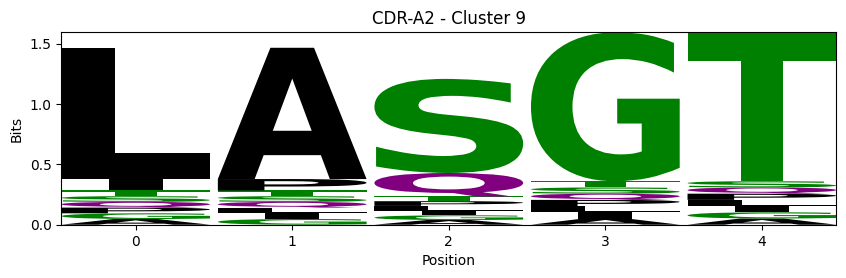
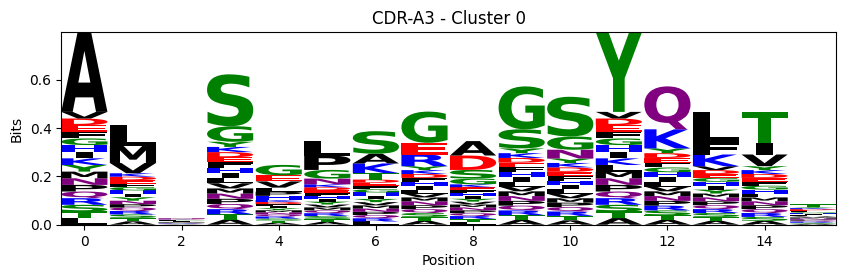
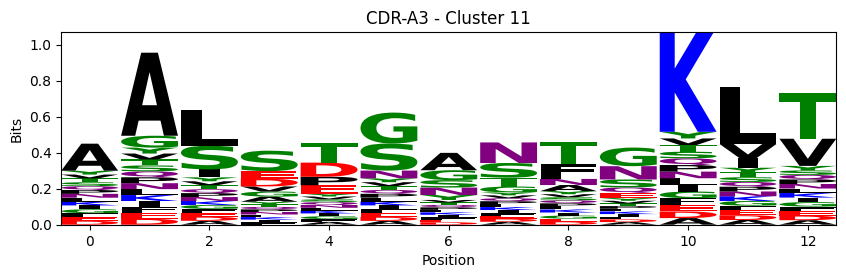
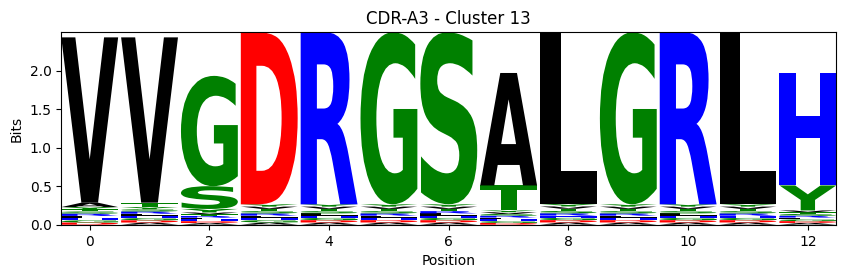
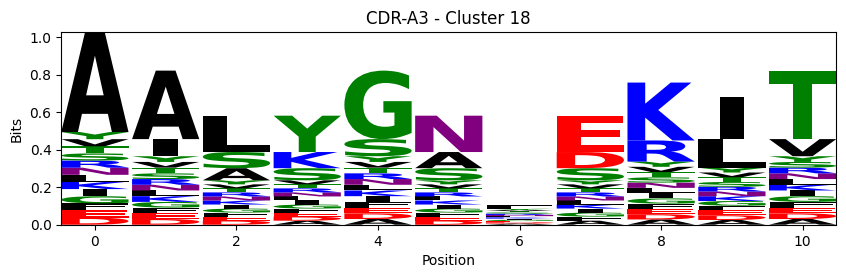
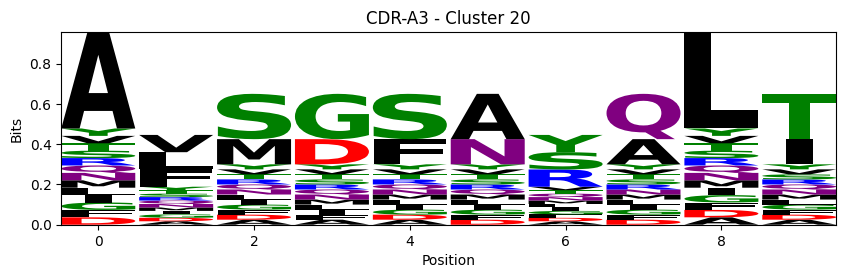
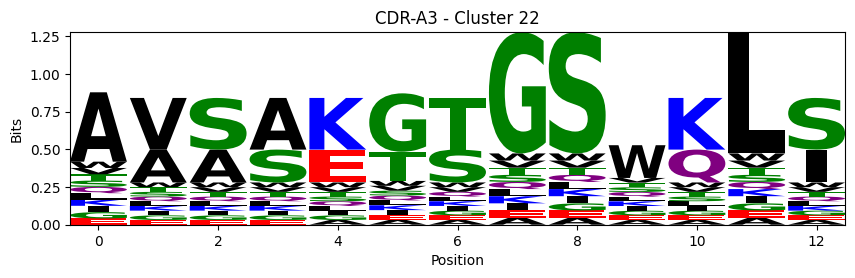
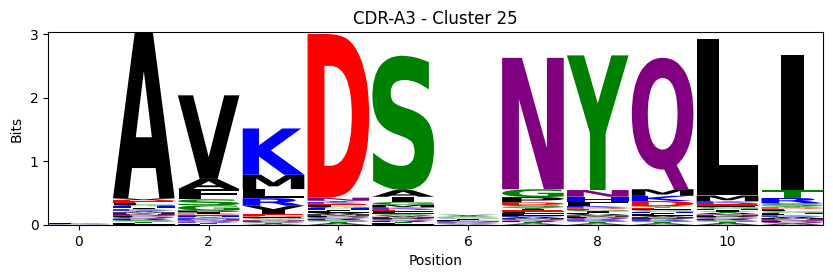
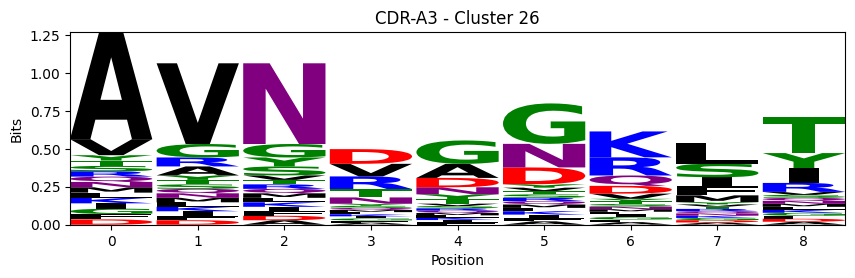
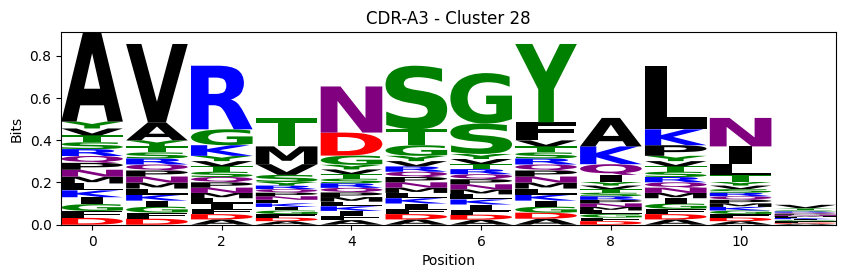
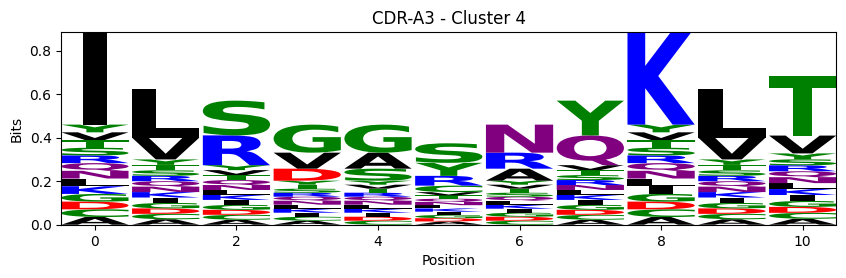
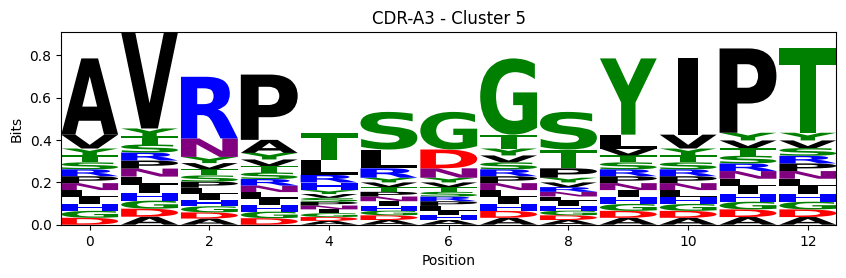
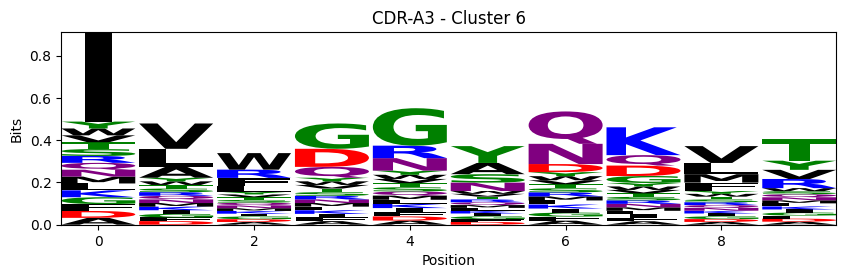
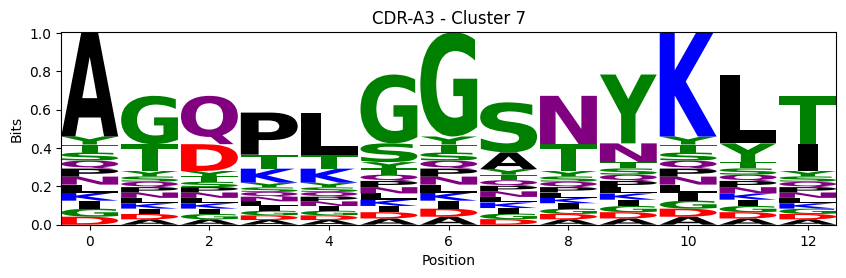
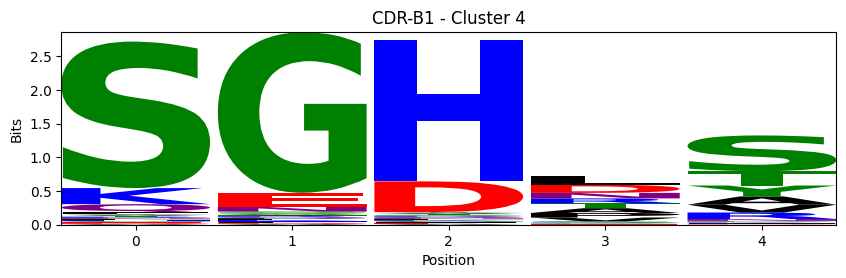
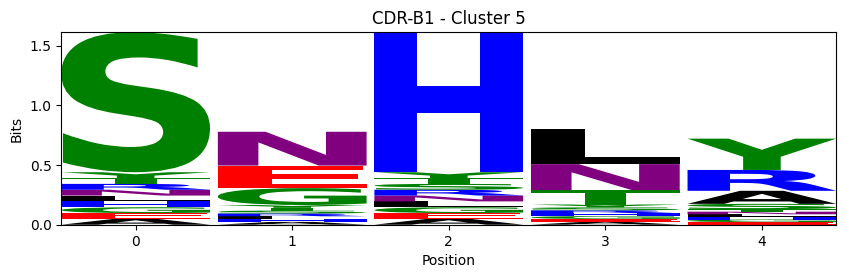
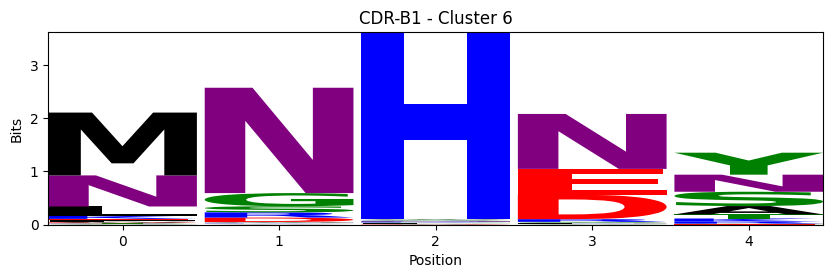
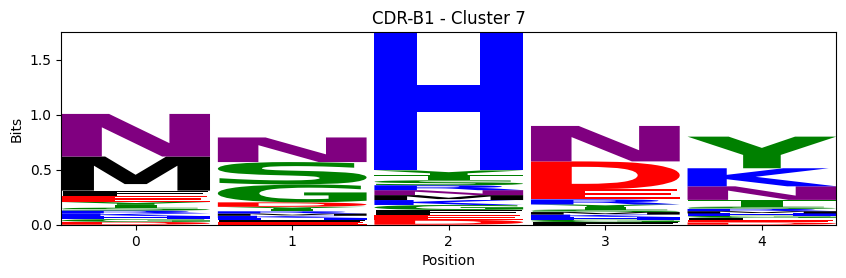
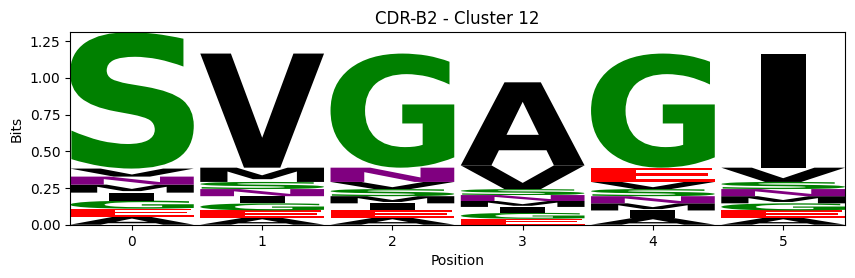
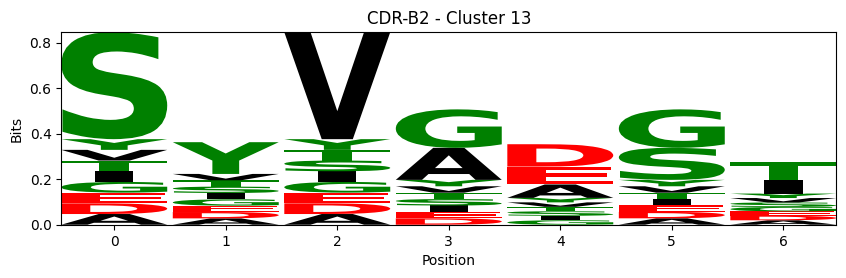
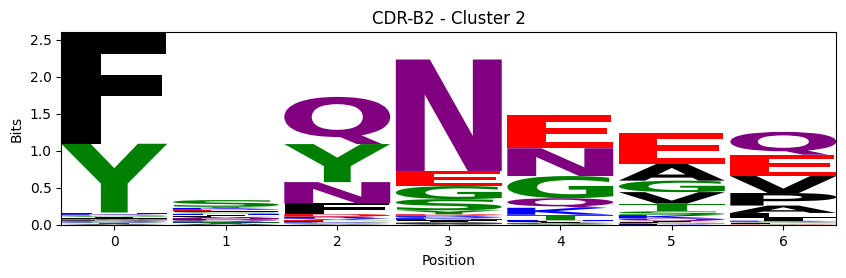
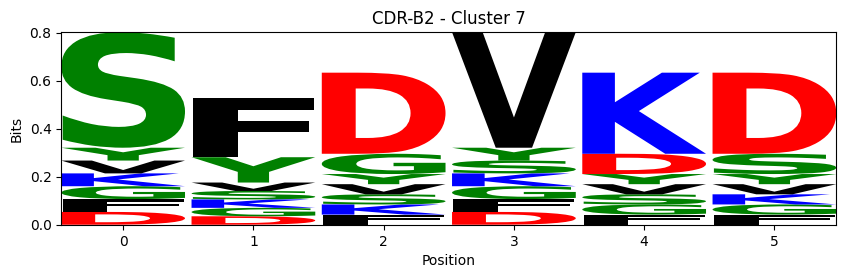
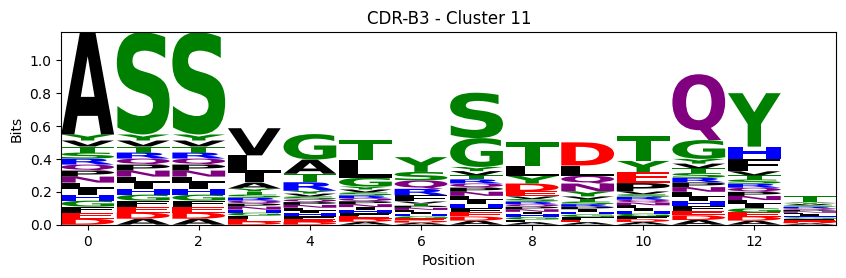
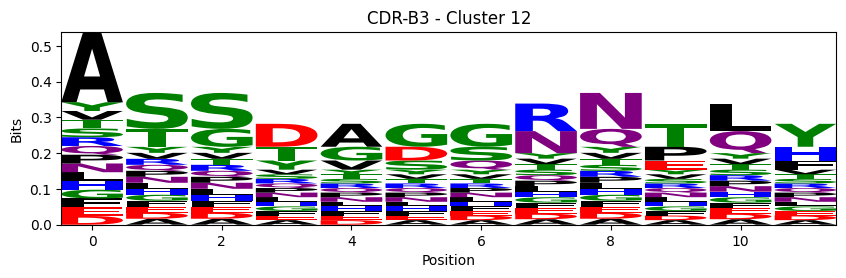
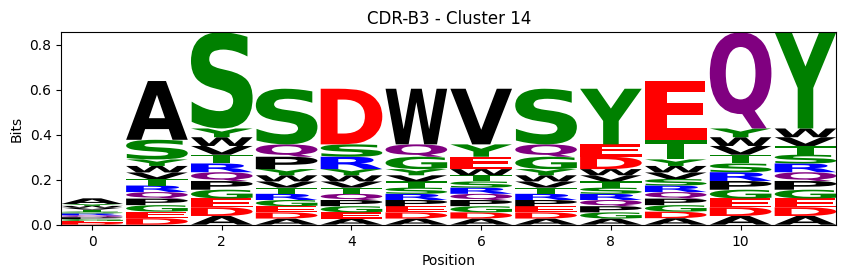
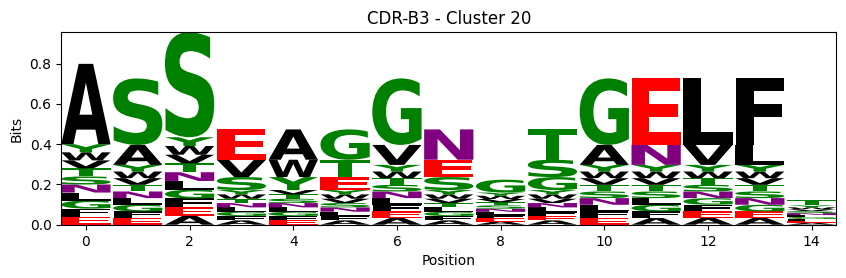
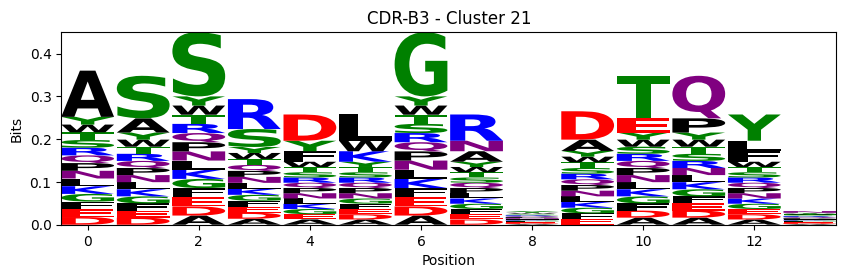
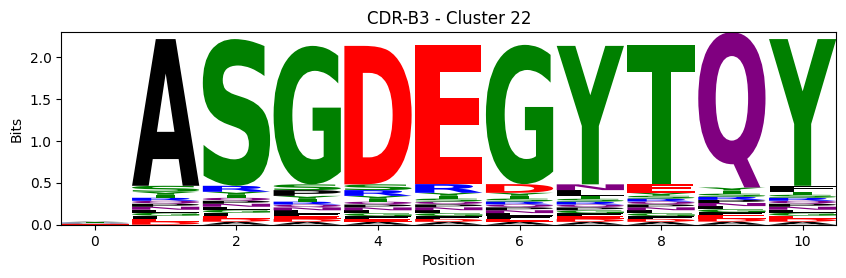
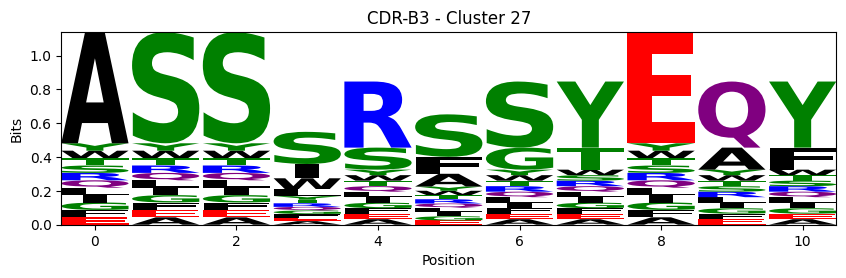
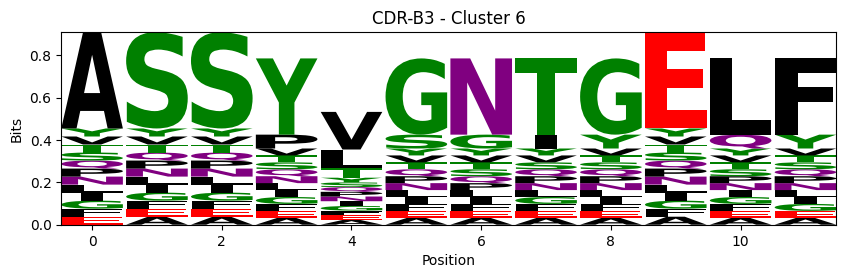
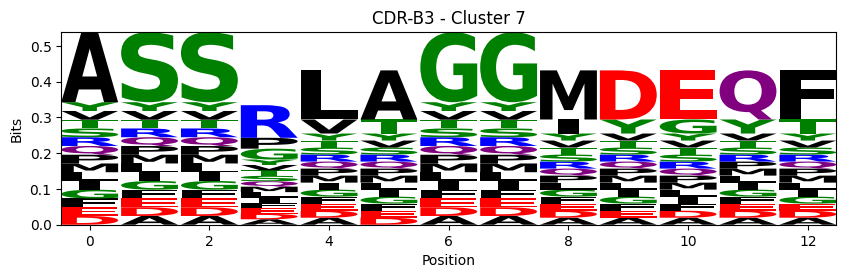
Sequence motifs for each canonical cluster
[11]:
def format_fasta(group):
index = group.index.tolist()
sequence = group['sequence'].tolist()
fasta = []
for idx, sequence in zip(index, sequence):
fasta.append(f'>{idx}\n{sequence}')
return '\n'.join(fasta)
[12]:
for (chain_type, cdr, cluster), group in (df.query("cluster != 'noise' and cluster_type == 'canonical'")
.groupby(['chain_type', 'cdr', 'cluster'])):
with tempfile.TemporaryDirectory() as directory:
input_file = os.path.join(directory, 'input.fasta')
output_file = os.path.join(directory, 'output.fasta')
with open(input_file, 'w') as fh:
fh.write(format_fasta(group))
cmd = f'clustalw2 -INFILE={input_file} -OUTFILE={output_file} -OUTPUT=FASTA'
subprocess.run(cmd.split(), check=True, stdout=subprocess.DEVNULL)
with open(output_file, 'r') as fh:
aligned_sequences = [line.strip() for line in fh.readlines() if not line.startswith('>')]
probs = logomaker.alignment_to_matrix(aligned_sequences, to_type='information')
logomaker.Logo(probs, color_scheme='chemistry')
cluster_name = f"CDR-{'A' if chain_type == 'alpha_chain' else 'B'}{cdr} - Cluster {cluster}"
plt.title(cluster_name)
plt.ylabel('Bits')
plt.xlabel('Position')
plt.show()
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
/project/koohylab/bmcmaste/miniconda3/envs/tcr-pmhc-interface-analysis/lib/python3.10/site-packages/logomaker/src/matrix.py:584: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
counts_df.loc[:, c] = tmp_mat.sum(axis=0).T
Look at backbone traces of each cluster
[13]:
ca_coordinates = []
names = []
chain_types = []
cdrs = []
clusters = []
for (chain_type, cdr, cluster), group in (df.query("cluster != 'noise' and cluster_type == 'canonical'")
.groupby(['chain_type', 'cdr', 'cluster'])):
pdb_id, chains = group.iloc[0]['name'].split('_')
alpha_chain_id, beta_chain_id = tuple(chains)
cdr = int(cdr)
with open(os.path.join('../data/raw/stcrdab', 'imgt', pdb_id + '.pdb'), 'r') as fh:
reference_structure_df = parse_pdb_to_pandas(fh.read())
reference_structure_df = annotate_tcr_pmhc_df(reference_structure_df, alpha_chain_id, beta_chain_id)
reference_tcr_df = reference_structure_df.query('chain_type.notnull()')
reference_cdr_df = reference_tcr_df.query(("chain_type == @chain_type and cdr == @cdr"))
refrence_anchors = pd.concat(find_anchors(reference_cdr_df, reference_tcr_df, 5))
for _, row in group.iterrows():
pdb_id, chains = row['name'].split('_')
alpha_chain_id, beta_chain_id = tuple(chains)
chain_type = row.chain_type
cdr = int(row.cdr)
with open(os.path.join('../data/raw/stcrdab', 'imgt', pdb_id + '.pdb'), 'r') as fh:
structure_df = parse_pdb_to_pandas(fh.read())
structure_df = annotate_tcr_pmhc_df(structure_df, alpha_chain_id, beta_chain_id)
tcr_df = structure_df.query('chain_type.notnull()')
cdr_df = tcr_df.query(("chain_type == @chain_type and cdr == @cdr"))
anchors = pd.concat(find_anchors(cdr_df, tcr_df, 5))
cdr_df = align_pandas_structure(
get_coords(anchors.query("atom_name == 'N' or atom_name == 'C' or atom_name == 'CA' or atom_name == 'O'")),
get_coords(refrence_anchors.query(("atom_name == 'N' or atom_name == 'C' "
"or atom_name == 'CA' or atom_name == 'O'"))),
cdr_df,
)
coords = get_coords(cdr_df.query("atom_name == 'CA'"))
ca_coordinates.append(coords)
chain_types.append(chain_type)
cdrs.append(cdr)
clusters.append(cluster)
names.append(row['name'])
ca_coordinate_df = pd.DataFrame({
'ca_coordinate': ca_coordinates,
'chain_type': chain_types,
'cdr': cdrs,
'name': names,
'cluster': clusters,
})
ca_coordinate_df = ca_coordinate_df.explode('ca_coordinate')
ca_coordinate_df[['ca_coordinate_x',
'ca_coordinate_y',
'ca_coordinate_z']] = ca_coordinate_df['ca_coordinate'].apply(pd.Series)
ca_coordinate_df = ca_coordinate_df.drop('ca_coordinate', axis=1)
ca_coordinate_df
[13]:
| chain_type | cdr | name | cluster | ca_coordinate_x | ca_coordinate_y | ca_coordinate_z | |
|---|---|---|---|---|---|---|---|
| 0 | alpha_chain | 1 | 4ozg_EF | 1 | 224.155000 | 40.712000 | 219.187000 |
| 0 | alpha_chain | 1 | 4ozg_EF | 1 | 227.900000 | 39.718000 | 219.665000 |
| 0 | alpha_chain | 1 | 4ozg_EF | 1 | 229.249000 | 39.147000 | 223.257000 |
| 0 | alpha_chain | 1 | 4ozg_EF | 1 | 232.636000 | 40.071000 | 224.817000 |
| 0 | alpha_chain | 1 | 4ozg_EF | 1 | 234.200000 | 36.568000 | 224.708000 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 2245 | beta_chain | 3 | 3gsn_AB | 7 | -55.580629 | 12.969075 | -2.638147 |
| 2245 | beta_chain | 3 | 3gsn_AB | 7 | -56.991148 | 13.552541 | -6.135106 |
| 2245 | beta_chain | 3 | 3gsn_AB | 7 | -55.012334 | 12.367820 | -9.171847 |
| 2245 | beta_chain | 3 | 3gsn_AB | 7 | -53.256231 | 14.847797 | -11.441973 |
| 2245 | beta_chain | 3 | 3gsn_AB | 7 | -54.906846 | 15.594755 | -14.752955 |
15537 rows × 7 columns
[14]:
ca_coordinate_df['cluster_name'] = (
'CDR-'
+ ca_coordinate_df['chain_type'].map(lambda chain_type: 'A' if chain_type == 'alpha_chain' else 'B')
+ ca_coordinate_df['cdr'].apply(str)
+ ' - Cluster '
+ ca_coordinate_df['cluster']
)
[15]:
for cluster_name, cluster_df in ca_coordinate_df.groupby('cluster_name'):
fig = px.line_3d()
cluster_df = ca_coordinate_df.query('cluster_name == @cluster_name')
for _, cdr_loop in cluster_df.groupby('name'):
fig.add_scatter3d(x=cdr_loop['ca_coordinate_x'],
y=cdr_loop['ca_coordinate_y'],
z=cdr_loop['ca_coordinate_z'],
mode='lines',
line=dict(color='green'),
showlegend=False)
fig.update_layout(title=cluster_name)
[16]:
def on_change(change):
if change['type'] == 'change' and change['name'] == 'value':
clear_output(wait=True)
dropdown = Dropdown(options=cluster_names, description='Select Cluster')
dropdown.observe(on_change)
display(VBox([dropdown]))
plot_cluster(change['new'])
def plot_cluster(cluster_name):
fig = px.line_3d()
cluster_df = ca_coordinate_df.query('cluster_name == @cluster_name')
for _, cdr_loop in cluster_df.groupby('name'):
fig.add_scatter3d(x=cdr_loop['ca_coordinate_x'],
y=cdr_loop['ca_coordinate_y'],
z=cdr_loop['ca_coordinate_z'],
mode='lines',
line=dict(color='green'),
showlegend=False)
fig.update_layout(title=cluster_name)
fig.show()
cluster_names = ca_coordinate_df['cluster_name'].unique().tolist()
dropdown = Dropdown(options=cluster_names, description='Select Cluster')
dropdown.observe(on_change)
display(VBox([dropdown]))
plot_cluster(cluster_names[0])